A first complete phylogenomic hypothesis for diploid blueberries (Vaccinium section Cyanococcus)
- PMID: 36109839
- PMCID: PMC10286767
- DOI: 10.1002/ajb2.16065
A first complete phylogenomic hypothesis for diploid blueberries (Vaccinium section Cyanococcus)
Abstract
Premise: The true blueberries (Vaccinium sect. Cyanococcus; Ericaceae), endemic to North America, have been intensively studied for over a century. However, with species estimates ranging from nine to 24 and much confusion regarding species boundaries, this ecologically and economically valuable group remains inadequately understood at a basic evolutionary and taxonomic level. As a first step toward understanding the evolutionary history and taxonomy of this species complex, we present the first phylogenomic hypothesis of the known diploid blueberries.
Methods: We used flow cytometry to verify the ploidy of putative diploid taxa and a target-enrichment approach to obtain a genomic data set for phylogenetic analyses.
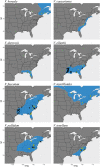
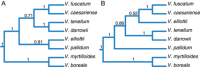
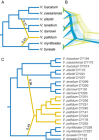
Results: Despite evidence of gene flow, we found that a primary phylogenetic signal is present. Monophyly for all morphospecies was recovered, with two notable exceptions: one sample of V. boreale was consistently nested in the V. myrtilloides clade and V. caesariense was nested in the V. fuscatum clade. One diploid taxon, Vaccinium pallidum, is implicated as having a homoploid hybrid origin.
Conclusions: This foundational study represents the first attempt to elucidate evolutionary relationships of the true blueberries of North America with a phylogenomic approach and sets the stage for multiple avenues of future study such as a taxonomic revision of the group, the verification of a homoploid hybrid taxon, and the study of polyploid lineages within the context of a diploid phylogeny.
Keywords: Ericaceae; HybSeq; Vaccinium; alleles; homoploid hybridization; phasing; phylogenetics; target enrichment.
© 2022 Botanical Society of America.
Figures

References
-
- Aalders, L. E. , and Hall I. V.. 1962. New evidence on the cytotaxonomy of Vaccinium species as revealed by stomatal measurements from herbarium specimens. Nature 196: 694. - PubMed
-
- Anderson, E. 1948. Hybridization of the habitat. Evolution 2: 1–9.
-
- Andrews, S. 2010. FastQC ‐ A quality control tool for high throughput sequence data. Website: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Arnold, M. L. 1993. Iris nelsonii (Iridaceae): Origin and genetic composition of a homoploid hybrid species. American Journal of Botany 80: 577–583. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

