Using ensembles and distillation to optimize the deployment of deep learning models for the classification of electronic cancer pathology reports
- PMID: 36110150
- PMCID: PMC9469924
- DOI: 10.1093/jamiaopen/ooac075
Using ensembles and distillation to optimize the deployment of deep learning models for the classification of electronic cancer pathology reports
Abstract
Objective: We aim to reduce overfitting and model overconfidence by distilling the knowledge of an ensemble of deep learning models into a single model for the classification of cancer pathology reports.
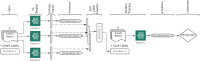
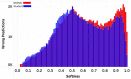
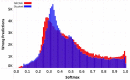
Materials and methods: We consider the text classification problem that involves 5 individual tasks. The baseline model consists of a multitask convolutional neural network (MtCNN), and the implemented ensemble (teacher) consists of 1000 MtCNNs. We performed knowledge transfer by training a single model (student) with soft labels derived through the aggregation of ensemble predictions. We evaluate performance based on accuracy and abstention rates by using softmax thresholding.
Results: The student model outperforms the baseline MtCNN in terms of abstention rates and accuracy, thereby allowing the model to be used with a larger volume of documents when deployed. The highest boost was observed for subsite and histology, for which the student model classified an additional 1.81% reports for subsite and 3.33% reports for histology.
Discussion: Ensemble predictions provide a useful strategy for quantifying the uncertainty inherent in labeled data and thereby enable the construction of soft labels with estimated probabilities for multiple classes for a given document. Training models with the derived soft labels reduce model confidence in difficult-to-classify documents, thereby leading to a reduction in the number of highly confident wrong predictions.
Conclusions: Ensemble model distillation is a simple tool to reduce model overconfidence in problems with extreme class imbalance and noisy datasets. These methods can facilitate the deployment of deep learning models in high-risk domains with low computational resources where minimizing inference time is required.
Keywords: CNN; NLP; deep learning; ensemble distillation; selective classification.
© The Author(s) 2022. Published by Oxford University Press on behalf of the American Medical Informatics Association.
Figures



References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A.. Cancer statistics, 2022. CA A Cancer J Clin 2022; 72 (1): 7–33. - PubMed
-
- Gao S, Qiu JX, Alawad M, et al.Classifying cancer pathology reports with hierarchical self-attention networks. Artif Intell Med 2019; 101: 101726. - PubMed
-
- Gao S, Ramanathan A, Tourassi G. Hierarchical convolutional attention networks for text classification. In: Proceedings of the Third Workshop on Representation Learning for NLP. Melbourne, Australia: Association for Computational Linguistics; 2018: 11–23. https://www.aclweb.org/anthology/W18-3002. Accessed September 1, 2022.
Grants and funding
LinkOut - more resources
Full Text Sources
