Classification and biomarker gene selection of pyroptosis-related gene expression in psoriasis using a random forest algorithm
- PMID: 36110207
- PMCID: PMC9468882
- DOI: 10.3389/fgene.2022.850108
Classification and biomarker gene selection of pyroptosis-related gene expression in psoriasis using a random forest algorithm
Abstract
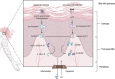
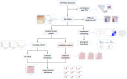
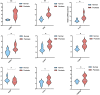
Background: Psoriasis is a chronic and immune-mediated skin disorder that currently has no cure. Pyroptosis has been proved to be involved in the pathogenesis and progression of psoriasis. However, the role pyroptosis plays in psoriasis remains elusive. Methods: RNA-sequencing data of psoriasis patients were obtained from the Gene Expression Omnibus (GEO) database, and differentially expressed pyroptosis-related genes (PRGs) between psoriasis patients and normal individuals were obtained. A principal component analysis (PCA) was conducted to determine whether PRGs could be used to distinguish the samples. PRG and immune cell correlation was also investigated. Subsequently, a novel diagnostic model comprising PRGs for psoriasis was constructed using a random forest algorithm (ntree = 400). A receiver operating characteristic (ROC) analysis was used to evaluate the classification performance through both internal and external validation. Consensus clustering analysis was used to investigate whether there was a difference in biological functions within PRG-based subtypes. Finally, the expression of the kernel PRGs were validated in vivo by qRT-PCR. Results: We identified a total of 39 PRGs, which could distinguish psoriasis samples from normal samples. The process of T cell CD4 memory activated and mast cells resting were correlated with PRGs. Ten PRGs, IL-1β, AIM2, CASP5, DHX9, CASP4, CYCS, CASP1, GZMB, CHMP2B, and CASP8, were subsequently screened using a random forest diagnostic model. ROC analysis revealed that our model has good diagnostic performance in both internal validation (area under the curve [AUC] = 0.930 [95% CI 0.877-0.984]) and external validation (mean AUC = 0.852). PRG subtypes indicated differences in metabolic processes and the MAPK signaling pathway. Finally, the qRT-PCR results demonstrated the apparent dysregulation of PRGs in psoriasis, especially AIM2 and GZMB. Conclusion: Pyroptosis may play a crucial role in psoriasis and could provide new insights into the diagnosis and underlying mechanisms of psoriasis.
Keywords: machine learning; psoriasis; pyroptosis; pyroptosis-related genes; random forest algorithm.
Copyright © 2022 Song, Zhang, Fei, Chen, Luo, Jiang, Ru, Xiang, Li, Luo and Kuai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Construction of the miRNA/Pyroptosis-Related Molecular Regulatory Axis in Abdominal Aortic Aneurysm: Evidence From Transcriptome Data Combined With Multiple Machine Learning Approaches Followed by Experiment Validation.J Immunol Res. 2024 Oct 30;2024:1429510. doi: 10.1155/2024/1429510. eCollection 2024. J Immunol Res. 2024. PMID: 39512836 Free PMC article.
-
Exploring hub pyroptosis-related genes, molecular subtypes, and potential drugs in ankylosing spondylitis by comprehensive bioinformatics analysis and molecular docking.BMC Musculoskelet Disord. 2023 Jun 29;24(1):532. doi: 10.1186/s12891-023-06664-8. BMC Musculoskelet Disord. 2023. PMID: 37386410 Free PMC article.
-
Deciphering the role of pyroptosis-related genes and natural killer T cells in sepsis pathogenesis: a comprehensive bioinformatics and Mendelian randomization analysis.J Physiol Pharmacol. 2025 Apr;76(2). doi: 10.26402/jpp.2025.2.10. Epub 2025 May 5. J Physiol Pharmacol. 2025. PMID: 40350654
-
Relevance of pyroptosis-associated genes in nasopharyngeal carcinoma diagnosis and subtype classification.J Gene Med. 2024 Jan;26(1):e3653. doi: 10.1002/jgm.3653. J Gene Med. 2024. PMID: 38282154
-
Pyroptosis-Related Genes as Diagnostic Markers in Chronic Obstructive Pulmonary Disease and Its Correlation with Immune Infiltration.Int J Chron Obstruct Pulmon Dis. 2024 Jun 27;19:1491-1513. doi: 10.2147/COPD.S438686. eCollection 2024. Int J Chron Obstruct Pulmon Dis. 2024. PMID: 38957709 Free PMC article.
Cited by
-
Metabolism-related biomarkers, molecular classification, and immune infiltration in diabetic ulcers with validation.Int Wound J. 2023 Nov;20(9):3498-3513. doi: 10.1111/iwj.14223. Epub 2023 May 28. Int Wound J. 2023. PMID: 37245869 Free PMC article.
-
Advancements and challenges of artificial intelligence in dermatology: a review of applications and perspectives in China.Front Digit Health. 2025 Aug 13;7:1544520. doi: 10.3389/fdgth.2025.1544520. eCollection 2025. Front Digit Health. 2025. PMID: 40881732 Free PMC article.
-
Advancing Psoriasis Care through Artificial Intelligence: A Comprehensive Review.Curr Dermatol Rep. 2024 Sep;13(3):141-147. doi: 10.1007/s13671-024-00434-y. Epub 2024 Jun 13. Curr Dermatol Rep. 2024. PMID: 39301276 Free PMC article.
-
Incorporating machine learning and PPI networks to identify mitochondrial fission-related immune markers in abdominal aortic aneurysms.Heliyon. 2024 Mar 26;10(7):e27989. doi: 10.1016/j.heliyon.2024.e27989. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38590878 Free PMC article.
-
CRISPR/Cas9-Mediated Knockout and Overexpression Studies Unveil the Role of PD-L1 in Immune Modulation in a Psoriasis-like Mouse Model.Inflammation. 2025 Apr 3. doi: 10.1007/s10753-025-02281-w. Online ahead of print. Inflammation. 2025. PMID: 40178656
References
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Royal Stat. Soc. Ser. B 57, 289–300. 10.1111/J.2517-6161.1995.TB02031.X - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

