RelB contributes to the survival, migration and lymphomagenesis of B cells with constitutively active CD40 signaling
- PMID: 36110848
- PMCID: PMC9468873
- DOI: 10.3389/fimmu.2022.913275
RelB contributes to the survival, migration and lymphomagenesis of B cells with constitutively active CD40 signaling
Abstract
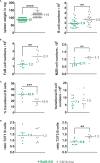
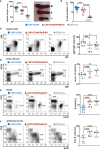
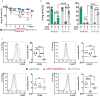
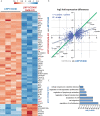
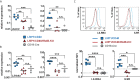
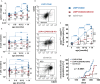
Activation of CD40-signaling contributes to the initiation, progression and drug resistance of B cell lymphomas. We contributed to this knowledge by showing that constitutive CD40-signaling in B cells induces B cell hyperplasia and finally B cell lymphoma development in transgenic mice. CD40 activates, among others, the non-canonical NF-ĸB signaling, which is constitutively activated in several human B cell lymphomas and is therefore presumed to contribute to lymphopathogenesis. This prompted us to study the regulatory role of the non-canonical NF-ĸB transcription factor RelB in lymphomagenesis. To this end, we crossed mice expressing a constitutively active CD40 receptor in B cells with conditional RelB-KO mice. Ablation of RelB attenuated pre-malignant B cell expansion, and resulted in an impaired survival and activation of long-term CD40-stimulated B cells. Furthermore, we found that hyperactivation of non-canonical NF-кB signaling enhances the retention of B cells in the follicles of secondary lymphoid organs. RNA-Seq-analysis revealed that several genes involved in B-cell migration, survival, proliferation and cytokine signaling govern the transcriptional differences modulated by the ablation of RelB in long-term CD40-stimulated B cells. Inactivation of RelB did not abrogate lymphoma development. However, lymphomas occurred with a lower incidence and had a longer latency period. In summary, our data suggest that RelB, although it is not strictly required for malignant transformation, accelerates the lymphomagenesis of long-term CD40-stimulated B cells by regulating genes involved in migration, survival and cytokine signaling.
Keywords: B cell lymphoma; CD40; IL9R; LILRB4; RelB; migration; non-canonical NF-ĸB-signaling; transgenic mice.
Copyright © 2022 Kuhn, Valentin, Stojanovic, Strobl, Babushku, Wang, Rambold, Scheffler, Grath, John-Robbert, Blum, Feuchtinger, Blutke, Weih, Kitamura, Rad, Strobl and Zimber-Strobl.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Constitutive CD40 signaling in B cells selectively activates the noncanonical NF-kappaB pathway and promotes lymphomagenesis.J Exp Med. 2008 Jun 9;205(6):1317-29. doi: 10.1084/jem.20080238. Epub 2008 May 19. J Exp Med. 2008. PMID: 18490492 Free PMC article.
-
CD40 ligand-mediated activation of the de novo RelB NF-kappaB synthesis pathway in transformed B cells promotes rescue from apoptosis.J Biol Chem. 2007 Jun 15;282(24):17475-85. doi: 10.1074/jbc.M607313200. Epub 2007 Apr 19. J Biol Chem. 2007. PMID: 17446175
-
CD40, but not lipopolysaccharide and anti-IgM stimulation of primary B lymphocytes, leads to a persistent nuclear accumulation of RelB.J Immunol. 1996 Dec 1;157(11):4862-9. J Immunol. 1996. PMID: 8943389
-
RelB, a member of the Rel/NF-kappa B family of transcription factors.Braz J Med Biol Res. 1996 Jul;29(7):895-903. Braz J Med Biol Res. 1996. PMID: 9070378 Review.
-
Effects of CD40 stimulation in the prevention of human EBV-lymphomagenesis.Leuk Lymphoma. 1997 Jan;24(3-4):187-99. doi: 10.3109/10428199709039007. Leuk Lymphoma. 1997. PMID: 9156649 Review.
Cited by
-
cFLIP in the molecular regulation of astroglia-driven neuroinflammation in experimental glaucoma.J Neuroinflammation. 2024 Jun 1;21(1):145. doi: 10.1186/s12974-024-03141-4. J Neuroinflammation. 2024. PMID: 38824526 Free PMC article.
-
The CD40/CD40L Pathway Regulates the Aggressiveness of Ovarian Cancer Cells via the Activation of Regulatory B Cells.Biochem Genet. 2024 Nov 22. doi: 10.1007/s10528-024-10945-9. Online ahead of print. Biochem Genet. 2024. PMID: 39576490
-
NF-κB fingerprinting reveals heterogeneous NF-κB composition in diffuse large B-cell lymphoma.Front Oncol. 2023 Jun 2;13:1181660. doi: 10.3389/fonc.2023.1181660. eCollection 2023. Front Oncol. 2023. PMID: 37333821 Free PMC article.
-
Single-nuclei transcriptome analysis of IgM+ cells isolated from channel catfish (Ictalurus punctatus) spleen.Front Immunol. 2025 Mar 17;16:1547193. doi: 10.3389/fimmu.2025.1547193. eCollection 2025. Front Immunol. 2025. PMID: 40165976 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

