A heterozygous p.S143P mutation in LMNA associates with proteasome dysfunction and enhanced autophagy-mediated degradation of mutant lamins A and C
- PMID: 36111332
- PMCID: PMC9468711
- DOI: 10.3389/fcell.2022.932983
A heterozygous p.S143P mutation in LMNA associates with proteasome dysfunction and enhanced autophagy-mediated degradation of mutant lamins A and C
Abstract
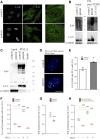
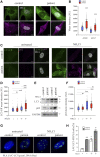
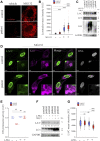
Lamins A and C are nuclear intermediate filament proteins that form a proteinaceous meshwork called lamina beneath the inner nuclear membrane. Mutations in the LMNA gene encoding lamins A and C cause a heterogenous group of inherited degenerative diseases known as laminopathies. Previous studies have revealed altered cell signaling pathways in lamin-mutant patient cells, but little is known about the fate of mutant lamins A and C within the cells. Here, we analyzed the turnover of lamins A and C in cells derived from a dilated cardiomyopathy patient with a heterozygous p.S143P mutation in LMNA. We found that transcriptional activation and mRNA levels of LMNA are increased in the primary patient fibroblasts, but the protein levels of lamins A and C remain equal in control and patient cells because of a meticulous interplay between autophagy and the ubiquitin-proteasome system (UPS). Both endogenous and ectopic expression of p.S143P lamins A and C cause significantly reduced activity of UPS and an accumulation of K48-ubiquitin chains in the nucleus. Furthermore, K48-ubiquitinated lamins A and C are degraded by compensatory enhanced autophagy, as shown by increased autophagosome formation and binding of lamins A and C to microtubule-associated protein 1A/1B-light chain 3. Finally, chaperone 4-PBA augmented protein degradation by restoring UPS activity as well as autophagy in the patient cells. In summary, our results suggest that the p.S143P-mutant lamins A and C have overloading and deleterious effects on protein degradation machinery and pharmacological interventions with compounds enhancing protein degradation may be beneficial for cell homeostasis.
Keywords: autophagy; degradation; disease mutations; lamin A/C (LMNA); ubiquitin (Ub); ubiquitin-proteasome degradation system.
Copyright © 2022 West, Turunen, Aalto, Virtanen, Li, Heliö, Meinander and Taimen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

