MLL regulates the actin cytoskeleton and cell migration by stabilising Rho GTPases via the expression of RhoGDI1
- PMID: 36111497
- PMCID: PMC7615853
- DOI: 10.1242/jcs.260042
MLL regulates the actin cytoskeleton and cell migration by stabilising Rho GTPases via the expression of RhoGDI1
Abstract
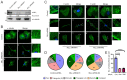
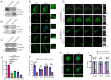
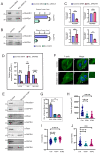
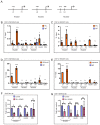
Attainment of proper cell shape and the regulation of cell migration are essential processes in the development of an organism. The mixed lineage leukemia (MLL or KMT2A) protein, a histone 3 lysine 4 (H3K4) methyltransferase, plays a critical role in cell-fate decisions during skeletal development and haematopoiesis in higher vertebrates. Rho GTPases - RhoA, Rac1 and CDC42 - are small G proteins that regulate various key cellular processes, such as actin cytoskeleton formation, the maintenance of cell shape and cell migration. Here, we report that MLL regulates the homeostasis of these small Rho GTPases. Loss of MLL resulted in an abnormal cell shape and a disrupted actin cytoskeleton, which lead to diminished cell spreading and migration. MLL depletion affected the stability and activity of Rho GTPases in a SET domain-dependent manner, but these Rho GTPases were not direct transcriptional targets of MLL. Instead, MLL regulated the transcript levels of their chaperone protein RhoGDI1 (also known as ARHGDIA). Using MDA-MB-231, a triple-negative breast cancer cell line with high RhoGDI1 expression, we show that MLL depletion or inhibition by small molecules reduces tumour progression in nude mice. Our studies highlight the central regulatory role of MLL in Rho/Rac/CDC42 signalling pathways. This article has an associated First Person interview with the first author of the paper.
Keywords: Cell migration; Cell shape; H3K4 methylation; MLL; Rho GTPases; RhoGDI1.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

