Identification of 2,4-Dinitro-Biphenyl-Based Compounds as MAPEG Inhibitors
- PMID: 36111583
- PMCID: PMC9827972
- DOI: 10.1002/cmdc.202200327
Identification of 2,4-Dinitro-Biphenyl-Based Compounds as MAPEG Inhibitors
Abstract
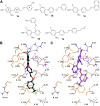
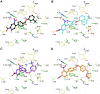
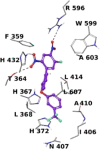
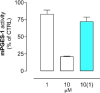
We identified 2,4-dinitro-biphenyl-based compounds as new inhibitors of leukotriene C4 synthase (LTC4 S) and 5-lipoxygenase-activating protein (FLAP), both members of the "Membrane Associated Proteins in Eicosanoid and Glutathione metabolism" (MAPEG) family involved in the biosynthesis of pro-inflammatory eicosanoids. By molecular docking we evaluated the putative binding against the targets of interest, and by applying cell-free and cell-based assays we assessed the inhibition of LTC4 S and FLAP by the small molecules at low micromolar concentrations. The present results integrate the previously observed inhibitory profile of the tested compounds against another MAPEG member, i. e., microsomal prostaglandin E2 synthase (mPGES)-1, suggesting that the 2,4-dinitro-biphenyl scaffold is a suitable molecular platform for a multitargeting approach to modulate pro-inflammatory mediators in inflammation and cancer treatment.
Keywords: FLAP inhibitors; LTC4S inhibitors; anti-inflammatory drugs; anticancer agents; multitargeting approach.
© 2022 The Authors. ChemMedChem published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Jakobsson P. J., Morgenstern R., Mancini J., Ford-Hutchinson A., Persson B., Am. J. Respir. Crit. Care Med. 2000, 161, S20–S24. - PubMed
-
- Liening S., Scriba G. K., Rummler S., Weinigel C., Kleinschmidt T. K., Haeggstrom J. Z., Werz O., Garscha U., Biochim. Biophys. Acta 2016, 1861, 1605–1613. - PubMed
-
- Thulasingam M., Haeggström J. Z., J. Mol. Biol. 2020, 432, 4999–5022. - PubMed
-
- Werz O., Gerstmeier J., Garscha U., Expert Opin. Ther. Pat. 2017, 27, 607–620. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

