An Extremes of Phenotype Approach Confirms Significant Genetic Heterogeneity in Patients with Ulcerative Colitis
- PMID: 36111848
- PMCID: PMC10024548
- DOI: 10.1093/ecco-jcc/jjac121
An Extremes of Phenotype Approach Confirms Significant Genetic Heterogeneity in Patients with Ulcerative Colitis
Abstract
Background and aims: Ulcerative colitis [UC] is a major form of inflammatory bowel disease globally. Phenotypic heterogeneity is defined by several variables including age of onset and disease extent. The genetics of disease severity remains poorly understood. To further investigate this, we performed a genome wide association [GWA] study using an extremes of phenotype strategy.
Methods: We conducted GWA analyses in 311 patients with medically refractory UC [MRUC], 287 with non-medically refractory UC [non-MRUC] and 583 controls. Odds ratios [ORs] were calculated for known risk variants comparing MRUC and non-MRUC, and controls.
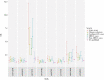
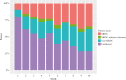
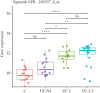
Results: MRUC-control analysis had the greatest yield of genome-wide significant single nucleotide polymorphisms [SNPs] [2018], including lead SNP = rs111838972 [OR = 1.82, p = 6.28 × 10-9] near MMEL1 and a locus in the human leukocyte antigen [HLA] region [lead SNP = rs144717024, OR = 12.23, p = 1.7 × 10-19]. ORs for the lead SNPs were significantly higher in MRUC compared to non-MRUC [p < 9.0 × 10-6]. No SNPs reached significance in the non-MRUC-control analysis (top SNP, rs7680780 [OR 2.70, p = 5.56 × 10-8). We replicate findings for rs4151651 in the Complement Factor B [CFB] gene and demonstrate significant changes in CFB gene expression in active UC. Detailed HLA analyses support the strong associations with MHC II genes, particularly HLA-DQA1, HLA-DQB1 and HLA-DRB1 in MRUC.
Conclusions: Our MRUC subgroup replicates multiple known UC risk variants in contrast to non-MRUC and demonstrates significant differences in effect sizes compared to those published. Non-MRUC cases demonstrate lower ORs similar to those published. Additional risk and prognostic loci may be identified by targeted recruitment of individuals with severe disease.
Keywords: Ulcerative colitis; disease severity; genetics.
© The Author(s) 2022. Published by Oxford University Press on behalf of European Crohn’s and Colitis Organisation.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures


References
-
- Solberg IC, Høivik ML, Cvancarova M, Moum B.. Risk matrix model for prediction of colectomy in a population-based study of ulcerative colitis patients (the IBSEN study). Scand J Gastroenterol 2015;50:1456–62. - PubMed
-
- Reinisch W, Reinink AR, Higgins PDR.. Factors associated with poor outcomes in adults with newly diagnosed ulcerative colitis. Clin Gastroenterol Hepatol 2015;13:635–42. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

