Epigenetic and pharmacological control of pigmentation via Bromodomain Protein 9 (BRD9)
- PMID: 36112085
- PMCID: PMC10091956
- DOI: 10.1111/pcmr.13068
Epigenetic and pharmacological control of pigmentation via Bromodomain Protein 9 (BRD9)
Abstract
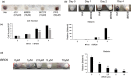
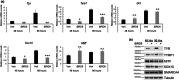
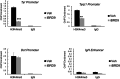
Lineage-specific differentiation programs are activated by epigenetic changes in chromatin structure. Melanin-producing melanocytes maintain a gene expression program ensuring appropriate enzymatic conversion of metabolites into the pigment, melanin, and transfer to surrounding cells. During neuroectodermal development, SMARCA4 (BRG1), the catalytic subunit of SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complexes, is essential for lineage specification. SMARCA4 is also required for development of multipotent neural crest precursors into melanoblasts, which differentiate into pigment-producing melanocytes. In addition to the catalytic domain, SMARCA4 and several SWI/SNF subunits contain bromodomains which are amenable to pharmacological inhibition. We investigated the effects of pharmacological inhibitors of SWI/SNF bromodomains on melanocyte differentiation. Strikingly, treatment of murine melanoblasts and human neonatal epidermal melanocytes with selected bromodomain inhibitors abrogated melanin synthesis and visible pigmentation. Using functional genomics, iBRD9, a small molecule selective for the bromodomain of BRD9 was found to repress pigmentation-specific gene expression. Depletion of BRD9 confirmed a requirement for expression of pigmentation genes in the differentiation program from melanoblasts into pigmented melanocytes and in melanoma cells. Chromatin immunoprecipitation assays showed that iBRD9 disrupts the occupancy of BRD9 and the catalytic subunit SMARCA4 at melanocyte-specific loci. These data indicate that BRD9 promotes melanocyte pigmentation whereas pharmacological inhibition of BRD9 is repressive.
Keywords: BRD9; SWI/SNF; bromodomain; chromatin remodeling; epigenetic; melanocyte differentiation; melanoma; pigmentation.
© 2022 The Authors. Pigment Cell & Melanoma Research published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures




Similar articles
-
BAF60A mediates interactions between the microphthalmia-associated transcription factor and the BRG1-containing SWI/SNF complex during melanocyte differentiation.J Cell Physiol. 2019 Jul;234(7):11780-11791. doi: 10.1002/jcp.27840. Epub 2018 Dec 4. J Cell Physiol. 2019. PMID: 30515787 Free PMC article.
-
Bromodomain and extra-terminal domain (BET) proteins regulate melanocyte differentiation.Epigenetics Chromatin. 2020 Mar 10;13(1):14. doi: 10.1186/s13072-020-00333-z. Epigenetics Chromatin. 2020. PMID: 32151278 Free PMC article.
-
BRG1 interacts with SOX10 to establish the melanocyte lineage and to promote differentiation.Nucleic Acids Res. 2017 Jun 20;45(11):6442-6458. doi: 10.1093/nar/gkx259. Nucleic Acids Res. 2017. PMID: 28431046 Free PMC article.
-
Epigenetic modulation by targeting bromodomain containing protein 9 (BRD9): Its therapeutic potential and selective inhibition.Int J Biol Macromol. 2023 Mar 1;230:123428. doi: 10.1016/j.ijbiomac.2023.123428. Epub 2023 Jan 26. Int J Biol Macromol. 2023. PMID: 36709803 Review.
-
SWI/SNF chromatin remodeling enzymes in melanocyte differentiation and melanoma.Crit Rev Eukaryot Gene Expr. 2014;24(2):151-61. doi: 10.1615/critreveukaryotgeneexpr.2014007882. Crit Rev Eukaryot Gene Expr. 2014. PMID: 24940768 Free PMC article. Review.
Cited by
-
Differential Contributions of mSWI/SNF Chromatin Remodeler Sub-Families to Myoblast Differentiation.Int J Mol Sci. 2023 Jul 9;24(14):11256. doi: 10.3390/ijms241411256. Int J Mol Sci. 2023. PMID: 37511016 Free PMC article.
-
Epigenetics-targeted drugs: current paradigms and future challenges.Signal Transduct Target Ther. 2024 Nov 26;9(1):332. doi: 10.1038/s41392-024-02039-0. Signal Transduct Target Ther. 2024. PMID: 39592582 Free PMC article. Review.
-
Targeting SWI/SNF Complexes in Cancer: Pharmacological Approaches and Implications.Epigenomes. 2024 Feb 4;8(1):7. doi: 10.3390/epigenomes8010007. Epigenomes. 2024. PMID: 38390898 Free PMC article. Review.
References
-
- Ahmed, N. S. , Gatchalian, J. , Ho, J. , Burns, M. J. , Hah, N. , Wei, Z. , Downes, M. , Evans, R. M. , & Hargreaves, D. C. (2022). BRD9 regulates interferon‐stimulated genes during macrophage activation via cooperation with BET protein BRD4. Proceedings of the National Academy of Sciences of the United States of America, 119(1), e2110812119. 10.1073/pnas.2110812119 - DOI - PMC - PubMed
-
- Alpsoy, A. , Utturkar, S. M. , Carter, B. C. , Dhiman, A. , Torregrosa‐Allen, S. E. , Currie, M. P. , Elzey, B. D. , & Dykhuizen, E. C. (2021). BRD9 is a critical regulator of androgen receptor signaling and prostate cancer progression. Cancer Research, 81(4), 820–833. 10.1158/0008-5472.CAN-20-1417 - DOI - PMC - PubMed
-
- Aras, S. , Saladi, S. V. , Basuroy, T. , Marathe, H. G. , Lores, P. , & de la Serna, I. L. (2019). BAF60A mediates interactions between the microphthalmia‐associated transcription factor and the BRG1‐containing SWI/SNF complex during melanocyte differentiation. Journal of Cellular Physiology, 234(7), 11780–11791. 10.1002/jcp.27840 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

