B-cell receptor physical properties affect relative IgG1 and IgE responses in mouse egg allergy
- PMID: 36114245
- PMCID: PMC9705252
- DOI: 10.1038/s41385-022-00567-y
B-cell receptor physical properties affect relative IgG1 and IgE responses in mouse egg allergy
Abstract
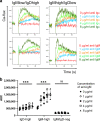
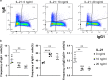
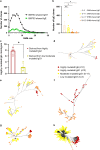
Mutated and unmutated IgE and IgG play different and partly opposing roles in allergy development, but the mechanisms controlling their relative production are incompletely understood. Here, we analyzed the IgE-response in murine food allergy. Deep sequencing of the complementary-determining region (CDR) repertoires indicated that an ongoing unmutated extrafollicular IgE response coexists with a germinal center response, even after long-lasting allergen challenges. Despite overall IgG1-dominance, a significant proportion of clonotypes contained several-fold more IgE than IgG1. Clonotypes with differential bias to either IgE or IgG1 showed distinct hypermutation and clonal expansion. Hypermutation rates were associated with different physiochemical binding properties of individual B-cell receptors (BCR). Increasing BCR signaling strength inhibited class switching from IgG1 to IgE in vitro, preferentially constraining IgE formation. These data indicate that antigen-binding properties of individual BCRs determine differential IgE hypermutation and IgE versus IgG1 production on the level of single B-cell clones.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

