The performance of genome sequencing as a first-tier test for neurodevelopmental disorders
- PMID: 36114283
- PMCID: PMC9822884
- DOI: 10.1038/s41431-022-01185-9
The performance of genome sequencing as a first-tier test for neurodevelopmental disorders
Abstract
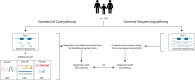
Genome sequencing (GS) can identify novel diagnoses for patients who remain undiagnosed after routine diagnostic procedures. We tested whether GS is a better first-tier genetic diagnostic test than current standard of care (SOC) by assessing the technical and clinical validity of GS for patients with neurodevelopmental disorders (NDD). We performed both GS and exome sequencing in 150 consecutive NDD patient-parent trios. The primary outcome was diagnostic yield, calculated from disease-causing variants affecting exonic sequence of known NDD genes. GS (30%, n = 45) and SOC (28.7%, n = 43) had similar diagnostic yield. All 43 conclusive diagnoses obtained with SOC testing were also identified by GS. SOC, however, required integration of multiple test results to obtain these diagnoses. GS yielded two more conclusive diagnoses, and four more possible diagnoses than ES-based SOC (35 vs. 31). Interestingly, these six variants detected only by GS were copy number variants (CNVs). Our data demonstrate the technical and clinical validity of GS to serve as routine first-tier genetic test for patients with NDD. Although the additional diagnostic yield from GS is limited, GS comprehensively identified all variants in a single experiment, suggesting that GS constitutes a more efficient genetic diagnostic workflow.
© 2022. The Author(s).
Conflict of interest statement
Part of the reagents required for the study were kindly provided by Illumina. However, Illumina was not involved in study design, the analyses and/or conclusions drawn from the data. The authors declare no conflict of interest.
Figures

Comment in
-
Genome sequencing as a single comprehensive test in molecular diagnosis.Eur J Hum Genet. 2023 Jan;31(1):3-4. doi: 10.1038/s41431-022-01215-6. Epub 2022 Oct 27. Eur J Hum Genet. 2023. PMID: 36289408 Free PMC article. No abstract available.
References
-
- Srivastava S, Love-Nichols JA, Dies KA, Ledbetter DH, Martin CL, Chung WK, et al. Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet Med. 2019;21:2413–21.. doi: 10.1038/s41436-019-0554-6. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

