Bud31-mediated alternative splicing is required for spermatogonial stem cell self-renewal and differentiation
- PMID: 36114296
- PMCID: PMC9883385
- DOI: 10.1038/s41418-022-01057-1
Bud31-mediated alternative splicing is required for spermatogonial stem cell self-renewal and differentiation
Abstract
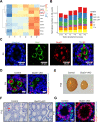
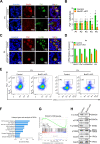
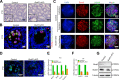
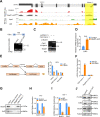
Alternative splicing (AS) is tightly regulated during cell differentiation and development. AS events are prevalent in the testis, but the splicing regulation in spermatogenesis remains unclear. Here we report that the spliceosome component Bud31 plays a crucial role during spermatogenesis in mice. Germ cell-specific knockout of Bud31 led to loss of spermatogonia and to male infertility. We further demonstrate that Bud31 is required for both spermatogonial stem cell pool maintenance and the initiation of spermatogenesis. SMART-seq revealed that deletion of Bud31 in germ cells causes widespread exon-skipping and intron retention. Particularly, we identified Cdk2 as one of the direct splicing targets of Bud31, knockout of Bud31 resulted in retention of the first intron of Cdk2, which led to a decrease in Cdk2 expression. Our findings suggest that Bud31-mediated AS within spermatogonial stem cells regulates the self-renewal and differentiation of male germ cells in mammals.
© 2022. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

