Individual competence predominates over host nutritional status in Arabidopsis root exudate-mediated bacterial enrichment in a combination of four Burkholderiaceae species
- PMID: 36114465
- PMCID: PMC9482264
- DOI: 10.1186/s12866-022-02633-8
Individual competence predominates over host nutritional status in Arabidopsis root exudate-mediated bacterial enrichment in a combination of four Burkholderiaceae species
Abstract
Background: Rhizosphere microorganisms play a crucial role in plant health and development. Plant root exudates (PRE) are a complex mixture of organic molecules and provide nutritional and signaling information to rhizosphere microorganisms. Burkholderiaceae species are non-abundant in the rhizosphere but exhibit a wide range of plant-growth-promoting and plant-health-protection effects. Most of these plant-associated microorganisms have been studied in isolation under laboratory conditions, whereas in nature, they interact in competition or cooperation with each other. To improve our understanding of the factors driving growth dynamics of low-abundant bacterial species in the rhizosphere, we hypothesized that the growth and survival of four Burkholderiaceae strains (Paraburkholderia phytofirmans PsJN, Cupriavidus metallidurans CH34, C. pinatubonensis JMP134 and C. taiwanensis LMG19424) in Arabidopsis thaliana PRE is affected by the presence of each other.
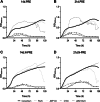
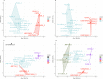
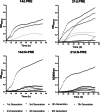
Results: Differential growth abilities of each strain were found depending on plant age and whether PRE was obtained after growth on N limitation conditions. The best-adapted strain to grow in PRE was P. phytofirmans PsJN, with C. pinatubonensis JMP134 growing better than the other two Cupriavidus strains. Individual strain behavior changed when they succeeded in combinations. Clustering analysis showed that the 4-member co-culture grouped with one of the best-adapted strains, either P. phytofirmans PsJN or C. pinatubonensis JMP134, depending on the PRE used. Sequential transference experiments showed that the behavior of the 4-member co-culture relies on the type of PRE provided for growth.
Conclusions: The results suggest that individual strain behavior changed when they grew in combinations of two, three, or four members, and those changes are determined first by the inherent characteristics of each strain and secondly by the environment.
Keywords: Arabidopsis; Bacterial growth; Burkholderiaceae; Co-culture; Root exudates.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

