Identification and validation of a gene-based signature reveals SLC25A10 as a novel prognostic indicator for patients with ovarian cancer
- PMID: 36114504
- PMCID: PMC9482156
- DOI: 10.1186/s13048-022-01039-4
Identification and validation of a gene-based signature reveals SLC25A10 as a novel prognostic indicator for patients with ovarian cancer
Abstract
Background: Ovarian cancer is a common gynecological cancer with poor prognosis and poses a serious threat to woman life and health. In this study, we aimed to establish a prognostic signature for the risk assessment of ovarian cancer.
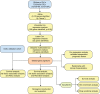
Methods: The Cancer Genome Atlas (TCGA) dataset was used as the training set and the International Cancer Genome Consortium (ICGC) dataset was set as an independent external validation. A multi-stage screening strategy was used to determine the prognostic features of ovarian cancer with R software. The relationship between the prognosis of ovarian cancer and the expression level of SLC25A10 was selected for further analysis.
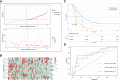
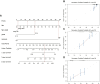
Results: A total of 16 prognosis-associated genes were screened to construct the risk score signature. Survival analysis showed that patients in the high-risk score group had a poor prognosis compared to the low-risk group. Accuracy of this prognostic signature was confirmed by the receiver operating characteristic (ROC) curve and decision curve analysis (DCA), and validated with ICGC cohort. This signature was identified as an independent factor for predicting overall survival (OS). Nomogram constructed by multiple clinical parameters showed excellent performance for OS prediction. Finally, it's found that patients with low expression of SLC25A10 generally had poor survival and higher resistance to most chemotherapeutic drugs.
Conclusions: In sum, we developed a 16-gene prognostic signature, which could serve as a promising tool for the prognostic prediction of ovarian cancer, and the expression level of SLC25A10 was tightly associated with OS of the patients.
Keywords: Ovarian Cancer; Prognosis; Risk Score; SLC25A10.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Identification and verification of a ten-gene signature predicting overall survival for ovarian cancer.Exp Cell Res. 2020 Oct 15;395(2):112235. doi: 10.1016/j.yexcr.2020.112235. Epub 2020 Aug 14. Exp Cell Res. 2020. PMID: 32805252
-
A prognostic model based on immune-related long noncoding RNAs for patients with epithelial ovarian cancer.J Ovarian Res. 2022 Jan 15;15(1):8. doi: 10.1186/s13048-021-00930-w. J Ovarian Res. 2022. PMID: 35031063 Free PMC article.
-
Identification and validation of a prognostic model based on immune-related genes in ovarian carcinoma.PeerJ. 2024 Oct 31;12:e18235. doi: 10.7717/peerj.18235. eCollection 2024. PeerJ. 2024. PMID: 39494284 Free PMC article.
-
Construction of a novel mRNA-signature prediction model for prognosis of bladder cancer based on a statistical analysis.BMC Cancer. 2021 Jul 27;21(1):858. doi: 10.1186/s12885-021-08611-z. BMC Cancer. 2021. PMID: 34315402 Free PMC article.
-
Novel immune-related gene signature for risk stratification and prognosis prediction in ovarian cancer.J Ovarian Res. 2023 Oct 19;16(1):205. doi: 10.1186/s13048-023-01289-w. J Ovarian Res. 2023. PMID: 37858138 Free PMC article.
Cited by
-
Targeting Mitochondrial Metabolic Reprogramming as a Potential Approach for Cancer Therapy.Int J Mol Sci. 2023 Mar 4;24(5):4954. doi: 10.3390/ijms24054954. Int J Mol Sci. 2023. PMID: 36902385 Free PMC article. Review.
-
An EMT-based gene signature enhances the clinical understanding and prognostic prediction of patients with ovarian cancers.J Ovarian Res. 2023 Mar 13;16(1):51. doi: 10.1186/s13048-023-01132-2. J Ovarian Res. 2023. PMID: 36907877 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

