Non-coding RNAs and epithelial mesenchymal transition in cancer: molecular mechanisms and clinical implications
- PMID: 36114510
- PMCID: PMC9479306
- DOI: 10.1186/s13046-022-02488-x
Non-coding RNAs and epithelial mesenchymal transition in cancer: molecular mechanisms and clinical implications
Abstract
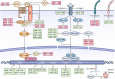
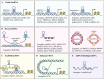
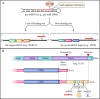
Epithelial-mesenchymal transition (EMT) is a fundamental process for embryonic development during which epithelial cells acquire mesenchymal characteristics, and the underlying mechanisms confer malignant features to carcinoma cells such as dissemination throughout the organism and resistance to anticancer treatments. During the past decades, an entire class of molecules, called non-coding RNA (ncRNA), has been characterized as a key regulator of almost every cellular process, including EMT. Like protein-coding genes, ncRNAs can be deregulated in cancer, acting as oncogenes or tumor suppressors. The various forms of ncRNAs, including microRNAs, PIWI-interacting RNAs, small nucleolar RNAs, transfer RNA-derived RNA fragments, long non-coding RNAs, and circular RNAs can orchestrate the complex regulatory networks of EMT at multiple levels. Understanding the molecular mechanism underlying ncRNAs in EMT can provide fundamental insights into cancer metastasis and may lead to novel therapeutic approaches. In this review, we describe recent advances in the understanding of ncRNAs in EMT and provide an overview of recent ncRNA applications in the clinic.
Keywords: Cancer; EMT; Metastasis; Molecular mechanisms; Non-coding RNA.
© 2022. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures
Similar articles
-
Non-coding RNAs regulating epithelial-mesenchymal transition: Research progress in liver disease.Biomed Pharmacother. 2022 Jun;150:112972. doi: 10.1016/j.biopha.2022.112972. Epub 2022 Apr 18. Biomed Pharmacother. 2022. PMID: 35447551 Review.
-
Genetic and epigenetic regulation of non-coding RNAs: Implications in cancer metastasis, stemness and drug resistance.Pathol Res Pract. 2025 Feb;266:155728. doi: 10.1016/j.prp.2024.155728. Epub 2024 Nov 28. Pathol Res Pract. 2025. PMID: 39657397 Review.
-
Non-coding RNAs and potential therapeutic targeting in cancer.Biochim Biophys Acta Rev Cancer. 2021 Jan;1875(1):188491. doi: 10.1016/j.bbcan.2020.188491. Epub 2020 Dec 13. Biochim Biophys Acta Rev Cancer. 2021. PMID: 33316377 Free PMC article. Review.
-
Non-coding RNAs as mediators of epithelial to mesenchymal transition in metastatic colorectal cancers.Cell Signal. 2025 Mar;127:111605. doi: 10.1016/j.cellsig.2025.111605. Epub 2025 Jan 20. Cell Signal. 2025. PMID: 39842529 Review.
-
Non-coding RNAs regulate tumor cell plasticity.Sci China Life Sci. 2013 Oct;56(10):886-90. doi: 10.1007/s11427-013-4554-5. Epub 2013 Oct 5. Sci China Life Sci. 2013. PMID: 24091685 Review.
Cited by
-
Co-Expression Network Analysis Unveiled lncRNA-mRNA Links Correlated to Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitor Resistance and/or Intermediate Epithelial-to-Mesenchymal Transition Phenotypes in a Human Non-Small Cell Lung Cancer Cellular Model System.Int J Mol Sci. 2024 Mar 29;25(7):3863. doi: 10.3390/ijms25073863. Int J Mol Sci. 2024. PMID: 38612674 Free PMC article.
-
The pivotal role of EMT-related noncoding RNAs regulatory axes in hepatocellular carcinoma.Front Pharmacol. 2023 Sep 11;14:1270425. doi: 10.3389/fphar.2023.1270425. eCollection 2023. Front Pharmacol. 2023. PMID: 37767397 Free PMC article. Review.
-
MALAT1 functions as a transcriptional promoter of MALAT1::GLI1 fusion for truncated GLI1 protein expression in cancer.BMC Cancer. 2023 May 10;23(1):424. doi: 10.1186/s12885-023-10867-6. BMC Cancer. 2023. PMID: 37165307 Free PMC article.
-
Interplay between lncRNA/miRNA and Wnt/ß-catenin signaling in brain cancer tumorigenesis.EXCLI J. 2023 Nov 28;22:1211-1222. doi: 10.17179/excli2023-6490. eCollection 2023. EXCLI J. 2023. PMID: 38204968 Free PMC article. Review.
-
Downregulation of N6-methyladenosine-modified LINC00641 promotes EMT, but provides a ferroptotic vulnerability in lung cancer.Cell Death Dis. 2023 Jun 13;14(6):359. doi: 10.1038/s41419-023-05880-3. Cell Death Dis. 2023. PMID: 37311754 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

