Replication-competent HIV-1 in human alveolar macrophages and monocytes despite nucleotide pools with elevated dUTP
- PMID: 36114511
- PMCID: PMC9482235
- DOI: 10.1186/s12977-022-00607-2
Replication-competent HIV-1 in human alveolar macrophages and monocytes despite nucleotide pools with elevated dUTP
Abstract
Background: Although CD4+ memory T cells are considered the primary latent reservoir for HIV-1, replication competent HIV has been detected in tissue macrophages in both animal and human studies. During in vitro HIV infection, the depleted nucleotide pool and high dUTP levels in monocyte derived macrophages (MDM) leads to proviruses with high levels of dUMP, which has been implicated in viral restriction or reduced transcription depending on the uracil base excision repair (UBER) competence of the macrophage. Incorporated dUMP has also been detected in viral DNA from circulating monocytes (MC) and alveolar macrophages (AM) of HIV infected patients on antiretroviral therapy (ART), establishing the biological relevance of this phenotype but not the replicative capacity of dUMP-containing proviruses.
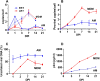
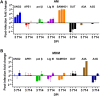
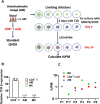
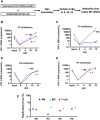
Results: As compared to in vitro differentiated MDM, AM from normal donors had sixfold lower levels of dTTP and a sixfold increased dUTP/dTTP, indicating a highly restrictive dNTP pool for reverse transcription. Expression of uracil DNA glycosylase (UNG) was eightfold lower in AM compared to the already low levels in MDM. Accordingly, ~ 80% of HIV proviruses contained dUMP, which persisted for at least 14-days due to low UNG excision activity. Unlike MDM, AM expression levels of UNG and SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 (SAMHD1) increased over 14 days post-HIV infection, while dUTP nucleotidohydrolase (DUT) expression decreased. These AM-specific effects suggest a restriction response centered on excising uracil from viral DNA copies and increasing relative dUTP levels. Despite the restrictive nucleotide pools, we detected rare replication competent HIV in AM, peripheral MC, and CD4+ T cells from ART-treated donors.
Conclusions: These findings indicate that the potential integration block of incorporated dUMP is not realized during in vivo infection of AM and MC due to the near absence of UBER activity. In addition, the increased expression of UNG and SAMHD1 in AM post-infection is too slow to prevent integration. Accordingly, dUMP persists in integrated viruses, which based on in vitro studies, can lead to transcriptional silencing. This possible silencing outcome of persistent dUMP could promote viral latency until the repressive effects of viral dUMP are reversed.
Keywords: Alveolar macrophage; Human HIV-1 infection; Monocyte; Nucleotide pools; Replication competent virus; Uracil base excision repair.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests
Figures

Similar articles
-
Deficient uracil base excision repair leads to persistent dUMP in HIV proviruses during infection of monocytes and macrophages.PLoS One. 2020 Jul 14;15(7):e0235012. doi: 10.1371/journal.pone.0235012. eCollection 2020. PLoS One. 2020. PMID: 32663205 Free PMC article.
-
Uracil DNA glycosylase initiates degradation of HIV-1 cDNA containing misincorporated dUTP and prevents viral integration.Proc Natl Acad Sci U S A. 2013 Feb 5;110(6):E448-57. doi: 10.1073/pnas.1219702110. Epub 2013 Jan 22. Proc Natl Acad Sci U S A. 2013. PMID: 23341616 Free PMC article.
-
Diverse fates of uracilated HIV-1 DNA during infection of myeloid lineage cells.Elife. 2016 Sep 20;5:e18447. doi: 10.7554/eLife.18447. Elife. 2016. PMID: 27644592 Free PMC article.
-
Uracils as a cellular weapon against viruses and mechanisms of viral escape.Curr HIV Res. 2006 Jan;4(1):31-42. doi: 10.2174/157016206775197673. Curr HIV Res. 2006. PMID: 16454709 Review.
-
SAMHD1 host restriction factor: a link with innate immune sensing of retrovirus infection.J Mol Biol. 2013 Dec 13;425(24):4981-94. doi: 10.1016/j.jmb.2013.10.022. Epub 2013 Oct 23. J Mol Biol. 2013. PMID: 24161438 Review.
Cited by
-
Chronic HIV Transcription, Translation, and Persistent Inflammation.Viruses. 2024 May 9;16(5):751. doi: 10.3390/v16050751. Viruses. 2024. PMID: 38793632 Free PMC article. Review.
-
HIV/Mtb Co-Infection: From the Amplification of Disease Pathogenesis to an "Emerging Syndemic".Microorganisms. 2023 Mar 27;11(4):853. doi: 10.3390/microorganisms11040853. Microorganisms. 2023. PMID: 37110276 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

