The role of DNA demethylation in liver to pancreas transdifferentiation
- PMID: 36114514
- PMCID: PMC9482206
- DOI: 10.1186/s13287-022-03159-6
The role of DNA demethylation in liver to pancreas transdifferentiation
Abstract
Background: Insulin producing cells generated by liver cell transdifferentiation, could serve as an attractive source for regenerative medicine. The present study assesses the relationship between DNA methylation pTFs induced liver to pancreas transdifferentiation.
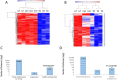
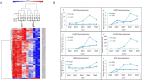
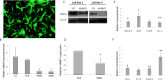
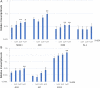
Results: The transdifferentiation process is associated with DNA demethylation, mainly at gene regulatory sites, and with increased expression of these genes. Active inhibition of DNA methylation promotes the pancreatic transcription factor-induced transdifferentiation process, supporting a causal role for DNA demethylation in this process.
Conclusions: Transdifferentiation is associated with global DNA hypomethylation, and with increased expression of specific demethylated genes. A combination of epigenetic modulators may be used to increase chromatin accessibility of the pancreatic transcription factors, thus promoting the efficiency of the developmental process.
Keywords: Cell replacement therapy for diabetes; DNA methylation; Epigenetic modifications; Liver; Pancreas; Pancreatic transcription factors; Transdifferentiation.
© 2022. The Author(s).
Conflict of interest statement
IML is an employee of Orgenesis LTD, SF is employee and consultant of Orgenesis LTD. Interpretation of results, and subsequent submission and publication decisions have been made independent of the sponsors. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Figures


References
-
- Meivar-Levy I, Sapir T, Gefen-Halevi S, Aviv V, Barshack I, Onaca N, et al. Pancreatic and duodenal homeobox gene 1 induces hepatic dedifferentiation by suppressing the expression of CCAAT/enhancer-binding protein beta. Hepatology (Baltimore, MD) 2007;46(3):898–905. doi: 10.1002/hep.21766. - DOI - PubMed
-
- Sapir T, Shternhall K, Meivar-Levy I, Blumenfeld T, Cohen H, Skutelsky E, et al. From the cover: cell-replacement therapy for diabetes: generating functional insulin-producing tissue from adult human liver cells. Proc Natl Acad Sci USA. 2005;102(22):7964–7969. doi: 10.1073/pnas.0405277102. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

