Enhancer RNA commits osteogenesis via microRNA-3129 expression in human bone marrow-derived mesenchymal stem cells
- PMID: 36114571
- PMCID: PMC9479228
- DOI: 10.1186/s41232-022-00228-4
Enhancer RNA commits osteogenesis via microRNA-3129 expression in human bone marrow-derived mesenchymal stem cells
Abstract
Background: Highly regulated gene expression program underlies osteogenesis of mesenchymal stem cells (MSCs), but the regulators in the program are not entirely identified. As enhancer RNAs (eRNAs) have recently emerged as a key regulator in gene expression, we assume a commitment of an eRNA in osteogenesis.
Methods: We performed in silico analysis to identify potential osteogenic microRNA (miRNA) gene predicted to be regulated by super-enhancers (SEs). SE inhibitor treatment and eRNA knocking-down were used to confirm the regulational mechanism of eRNA. miRNA function in osteogenesis was elucidated by miR mimic and inhibitor transfection experiments.
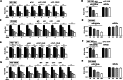
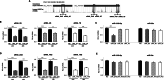
Results: miR-3129 was found to be located adjacent in a SE (osteoblast-specific SE_46171) specifically activated in osteoblasts by in silico analysis. A RT-quantitative PCR analysis of human bone marrow-derived MSC (hBMSC) cells showed that eRNA_2S was transcribed from the SE with the expression of miR-3129. Knockdown of eRNA_2S by locked nucleic acid as well as treatment of SE inhibitors JQ1 or THZ1 resulted in low miR-3129 levels. Overexpression of miR-3129 promoted hBMSC osteogenesis, while knockdown of miR-3129 inhibited hBMSC osteogenesis. Solute carrier family 7 member 11 (SLC7A11), encoding a bone formation suppressor, was upregulated following miR-3129-5p inhibition and identified as a target gene for miR-3129 during differentiation of hBMSCs into osteoblasts.
Conclusions: miR-3129 expression is regulated by SEs via eRNA_2S and this miRNA promotes hBMSC differentiation into osteoblasts through downregulating the target gene SLC7A11. Thus, the present study uncovers a commitment of an eRNA via a miR-3129/SLC7A11 regulatory pathway during osteogenesis of hBMSCs.
Keywords: Enhancer RNA; Mesenchymal stem cells; Osteogenesis; Super-enhancer; miR-3129.
© 2022. The Author(s).
Conflict of interest statement
Y. Tanaka has received consulting fees, speaking fees, and/or honoraria from Daiichi-Sankyo, Astellas, Pfizer, Mitsubishi-Tanabe, Bristol-Myers, Chugai, YL Biologics, Eli Lilly, Sanofi, Janssen, and UCB and has received research grants from Mitsubishi-Tanabe, Takeda, Bristol-Myers, Chugai, Astellas, Abbvie, MSD, Daiichi-Sankyo, Pfizer, Kyowa-Kirin, Eisai, and Ono. S. Nakayamada has received consulting fees, speaking fees, and/or honoraria from Bristol-Myers, AstraZeneca, Pfizer, GlaxoSmithKline, Astellas, Asahi-kasei, Sanofi, Abbvie, Eisai, Chugai, Gilead, Boehringer Ingelheim and has received research grants from Mitsubishi-Tanabe.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials

