Targeted genomic translocations and inversions generated using a paired prime editing strategy
- PMID: 36114670
- PMCID: PMC9840113
- DOI: 10.1016/j.ymthe.2022.09.008
Targeted genomic translocations and inversions generated using a paired prime editing strategy
Abstract
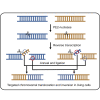
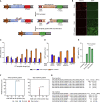
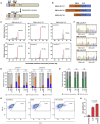
A variety of cancers have been found to have chromosomal rearrangements, and the genomic abnormalities often induced expression of fusion oncogenes. To date, a pair of engineered nucleases including ZFNs, TALENs, and CRISPR-Cas9 nucleases have been used to generate chromosomal rearrangement in living cells and organisms for disease modeling. However, these methods induce unwanted indel mutations at the DNA break junctions, resulting in incomplete disease modeling. Here, we developed prime editor nuclease-mediated translocation and inversion (PETI), a method for programmable chromosomal translocation and inversion using prime editor 2 nuclease (PE2 nuclease) and paired pegRNA. Using PETI method, we successfully introduced DNA recombination in episomal fluorescence reporters as well as precise chromosomal translocations in human cells. We applied PETI to create cancer-associated translocations and inversions such as NPM1-ALK and EML4-ALK in human cells. Our findings show that PETI generated chromosomal translocation and inversion in a programmable manner with efficiencies comparable of Cas9. PETI methods, we believe, could be used to create disease models or for gene therapy.
Keywords: CRISPR-Cas9; chromosomal inversion; chromosomal translocation; genome editing; genome rearrangement; prime editing.
Copyright © 2022 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare that they have no competing interests.
Figures


Similar articles
-
Detection and Modulation of DNA Translocations During Multi-Gene Genome Editing in T Cells.CRISPR J. 2020 Jun;3(3):177-187. doi: 10.1089/crispr.2019.0074. CRISPR J. 2020. PMID: 32584143
-
Genome Editing of Structural Variations: Modeling and Gene Correction.Trends Biotechnol. 2016 Jul;34(7):548-561. doi: 10.1016/j.tibtech.2016.02.011. Epub 2016 Mar 23. Trends Biotechnol. 2016. PMID: 27016031 Review.
-
Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions.Genetics. 2013 Oct;195(2):331-48. doi: 10.1534/genetics.113.155382. Epub 2013 Aug 9. Genetics. 2013. PMID: 23934893 Free PMC article.
-
Induction of Chromosomal Translocations with CRISPR-Cas9 and Other Nucleases: Understanding the Repair Mechanisms That Give Rise to Translocations.Adv Exp Med Biol. 2018;1044:15-25. doi: 10.1007/978-981-13-0593-1_2. Adv Exp Med Biol. 2018. PMID: 29956288 Free PMC article. Review.
-
Prime editing in plants and mammalian cells: Mechanism, achievements, limitations, and future prospects.Bioessays. 2022 Sep;44(9):e2200032. doi: 10.1002/bies.202200032. Epub 2022 Jun 24. Bioessays. 2022. PMID: 35750651 Review.
Cited by
-
Deconstructing cancer with precision genome editing.Biochem Soc Trans. 2024 Apr 24;52(2):803-819. doi: 10.1042/BST20230984. Biochem Soc Trans. 2024. PMID: 38629716 Free PMC article. Review.
-
Recent advances in prime editing technologies and their promises for therapeutic applications.Curr Opin Biotechnol. 2024 Apr;86:103071. doi: 10.1016/j.copbio.2024.103071. Epub 2024 Feb 7. Curr Opin Biotechnol. 2024. PMID: 38330875 Free PMC article. Review.
-
Dual inhibition of DNA-PK and Polϴ boosts precision of diverse prime editing systems.Nat Commun. 2025 May 8;16(1):4290. doi: 10.1038/s41467-025-59708-z. Nat Commun. 2025. PMID: 40341582 Free PMC article.
-
Integrating Prime Editing and Cellular Reprogramming as Novel Strategies for Genetic Cardiac Disease Modeling and Treatment.Curr Cardiol Rep. 2024 Nov;26(11):1197-1208. doi: 10.1007/s11886-024-02118-2. Epub 2024 Sep 11. Curr Cardiol Rep. 2024. PMID: 39259489 Free PMC article. Review.
-
Advancing Precision Medicine: Recent Innovations in Gene Editing Technologies.Adv Sci (Weinh). 2025 Apr;12(14):e2410237. doi: 10.1002/advs.202410237. Epub 2025 Mar 2. Adv Sci (Weinh). 2025. PMID: 40025867 Free PMC article. Review.
References
-
- Stankiewicz P., Lupski J.R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010;61:437–455. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

