PFKFB4 interacts with FBXO28 to promote HIF-1α signaling in glioblastoma
- PMID: 36115843
- PMCID: PMC9482633
- DOI: 10.1038/s41389-022-00433-3
PFKFB4 interacts with FBXO28 to promote HIF-1α signaling in glioblastoma
Abstract
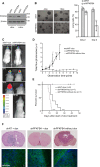
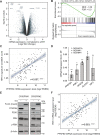
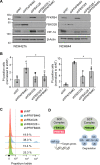
Glioblastoma is a highly aggressive brain tumor for which there is no cure. The metabolic enzyme 6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase 4 (PFKFB4) is essential for glioblastoma stem-like cell (GSC) survival but its mode of action is unclear. Understanding the role of PFKFB4 in tumor cell survival could allow it to be leveraged in a cancer therapy. Here, we show the importance of PFKFB4 for glioblastoma growth in vivo in an orthotopic patient derived mouse model. In an evaluation of patient tumor samples of different cancer entities, PFKFB4 protein was found to be overexpressed in prostate, lung, colon, mammary and squamous cell carcinoma, with expression level correlating with tumor grade. Gene expression profiling in PFKFB4-silenced GSCs revealed a downregulation of hypoxia related genes and Western blot analysis confirmed a dramatic reduction of HIF (hypoxia inducible factor) protein levels. Through mass spectrometric analysis of immunoprecipitated PFKFB4, we identified the ubiquitin E3 ligase, F-box only protein 28 (FBXO28), as a new interaction partner of PFKFB4. We show that PFKFB4 regulates the ubiquitylation and subsequent proteasomal degradation of HIF-1α, which is mediated by the ubiquitin ligase activity of FBXO28. This newly discovered function of PFKFB4, coupled with its cancer specificity, provides a new strategy for inhibiting HIF-1α in cancer cells.
© 2022. The Author(s).
Conflict of interest statement
DKFZ filed the patent application EP19701524.1 based on the results of this study and VG, EP, JB ad FB are listed as inventor. The authors declare no conflict of interest.
Figures



References
-
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–8. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

