De novo annotation of lncRNA HOTAIR transcripts by long-read RNA capture-seq reveals a differentiation-driven isoform switch
- PMID: 36115964
- PMCID: PMC9482196
- DOI: 10.1186/s12864-022-08887-w
De novo annotation of lncRNA HOTAIR transcripts by long-read RNA capture-seq reveals a differentiation-driven isoform switch
Abstract
Background: LncRNAs are tissue-specific and emerge as important regulators of various biological processes and as disease biomarkers. HOTAIR is a well-established pro-oncogenic lncRNA which has been attributed a variety of functions in cancer and native contexts. However, a lack of an exhaustive, cell type-specific annotation questions whether HOTAIR functions are supported by the expression of multiple isoforms.
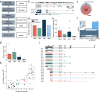
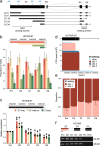
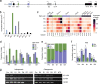
Results: Using a capture long-read sequencing approach, we characterize HOTAIR isoforms expressed in human primary adipose stem cells. We find HOTAIR isoforms population displays varied splicing patterns, frequently leading to the exclusion or truncation of canonical LSD1 and PRC2 binding domains. We identify a highly cell type-specific HOTAIR isoform pool regulated by distinct promoter usage, and uncover a shift in the HOTAIR TSS usage that modulates the balance of HOTAIR isoforms at differentiation onset.
Conclusion: Our results highlight the complexity and cell type-specificity of HOTAIR isoforms and open perspectives on functional implications of these variants and their balance to key cellular processes.
Keywords: Adipose differentiation; Adipose stem cells; Capture-seq; HOTAIR; Long-read sequencing; lncRNA isoform.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
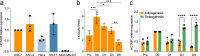
Similar articles
-
Global profiling of hnRNP A2/B1-RNA binding on chromatin highlights LncRNA interactions.RNA Biol. 2018;15(7):901-913. doi: 10.1080/15476286.2018.1474072. Epub 2018 Jul 25. RNA Biol. 2018. PMID: 29938567 Free PMC article.
-
HOTAIR lncRNA promotes epithelial-mesenchymal transition by redistributing LSD1 at regulatory chromatin regions.EMBO Rep. 2021 Jul 5;22(7):e50193. doi: 10.15252/embr.202050193. Epub 2021 May 6. EMBO Rep. 2021. PMID: 33960111 Free PMC article.
-
JMJD6-BRD4 complex stimulates lncRNA HOTAIR transcription by binding to the promoter region of HOTAIR and induces radioresistance in liver cancer stem cells.J Transl Med. 2023 Oct 25;21(1):752. doi: 10.1186/s12967-023-04394-y. J Transl Med. 2023. PMID: 37880710 Free PMC article.
-
HOTAIR: An Oncogenic Long Non-Coding RNA in Human Cancer.Cell Physiol Biochem. 2018;47(3):893-913. doi: 10.1159/000490131. Epub 2018 May 24. Cell Physiol Biochem. 2018. PMID: 29843138 Review.
-
LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer.Biochim Biophys Acta. 2015 Aug;1856(1):151-64. doi: 10.1016/j.bbcan.2015.07.001. Epub 2015 Jul 21. Biochim Biophys Acta. 2015. PMID: 26208723 Free PMC article. Review.
Cited by
-
TMEM244 Is a Long Non-Coding RNA Necessary for CTCL Cell Growth.Int J Mol Sci. 2023 Feb 9;24(4):3531. doi: 10.3390/ijms24043531. Int J Mol Sci. 2023. PMID: 36834942 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

