Prediction of Functional Genes in Primary Varicose Great Saphenous Veins Using the lncRNA-miRNA-mRNA Network
- PMID: 36118829
- PMCID: PMC9477642
- DOI: 10.1155/2022/4722483
Prediction of Functional Genes in Primary Varicose Great Saphenous Veins Using the lncRNA-miRNA-mRNA Network
Retraction in
-
Retracted: Prediction of Functional Genes in Primary Varicose Great Saphenous Veins Using the lncRNA-miRNA-mRNA Network.Comput Math Methods Med. 2023 Nov 29;2023:9769684. doi: 10.1155/2023/9769684. eCollection 2023. Comput Math Methods Med. 2023. PMID: 38077881 Free PMC article.
Abstract
Background: Long noncoding RNAs (lncRNAs) have been widely suggested to bind with the microRNA (miRNA) sites and play roles of competing endogenous RNAs (ceRNAs), which can thus affect and regulate target gene and mRNA expression. Such lncRNA-related ceRNAs are identified to exert vital parts in vascular disease. Nonetheless, it remains unknown about how the lncRNA-miRNA-mRNA network functions in the varicose great saphenous veins.
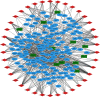
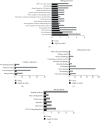
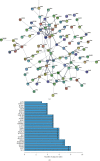
Methods: This study acquired the lncRNA and mRNA expression patterns from the GEO database and identifies the differentially expressed mRNAs and lncRNAs by adopting the R software "limma" package. Then, miRcode, miRDB, miRTarbase, and TargetScan were used to establish the miRNA-mRNA pairs and lncRNA-miRNA pairs. In addition, the lncRNA-miRNA-mRNA ceRNA network was constructed by using Cytoscape. Protein-protein interaction, Gene Ontology functional annotations, and Kyoto Encyclopedia of Genes and Genomes enrichment were carried out to examine the candidate hub genes, the functions of genes, and the corresponding pathways.
Results: In line with the preset theory, we constructed ceRNA network comprising 12 lncRNAs, 38 miRNAs, and 149 mRNAs. Kyoto Encyclopedia of Genes and Genomes analysis indicated that the PI3K/Akt signaling pathway played a vital part in the development of varicose great saphenous veins. AC114730, AC002127, and AC073342 were significant biomarkers. At the same time, we predicted the potential miRNA, which may exert a significant influence on the varicose great saphenous veins, namely, miR-17-5p, miR-129-5p, miR-1297, miR-20b-5p, and miR-33a-3p.
Conclusion: By performing ceRNA network analysis, our study detects new lncRNAs, miRNAs, and mRNAs, which can be applied as underlying biomarkers of varicose great saphenous veins and as therapeutic targets for the treatment of varicose great saphenous veins.
Copyright © 2022 Jiabo Wei et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Comprehensive analysis of lncRNA-miRNA-mRNA networks during osteogenic differentiation of bone marrow mesenchymal stem cells.BMC Genomics. 2022 Jun 7;23(1):425. doi: 10.1186/s12864-022-08646-x. BMC Genomics. 2022. PMID: 35672672 Free PMC article.
-
Identification of a lncRNA/circRNA-miRNA-mRNA ceRNA Network in Alzheimer's Disease.J Integr Neurosci. 2023 Oct 17;22(6):136. doi: 10.31083/j.jin2206136. J Integr Neurosci. 2023. PMID: 38176923
-
Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue.BMC Cancer. 2019 Aug 7;19(1):779. doi: 10.1186/s12885-019-5983-8. BMC Cancer. 2019. PMID: 31391008 Free PMC article.
-
Reconstruction and Analysis of the Differentially Expressed IncRNA-miRNA-mRNA Network Based on Competitive Endogenous RNA in Hepatocellular Carcinoma.Crit Rev Eukaryot Gene Expr. 2019;29(6):539-549. doi: 10.1615/CritRevEukaryotGeneExpr.2019028740. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422009 Review.
-
Bioinformatics analysis of ceRNA network of autophagy-related genes in pediatric asthma.Medicine (Baltimore). 2023 Dec 1;102(48):e36343. doi: 10.1097/MD.0000000000036343. Medicine (Baltimore). 2023. PMID: 38050261 Free PMC article. Review.
Cited by
-
Identification of biological significance of different stages of varicose vein development based on mRNA sequencing.Sci Rep. 2024 Sep 28;14(1):22536. doi: 10.1038/s41598-024-73691-3. Sci Rep. 2024. PMID: 39341975 Free PMC article.
-
Retracted: Prediction of Functional Genes in Primary Varicose Great Saphenous Veins Using the lncRNA-miRNA-mRNA Network.Comput Math Methods Med. 2023 Nov 29;2023:9769684. doi: 10.1155/2023/9769684. eCollection 2023. Comput Math Methods Med. 2023. PMID: 38077881 Free PMC article.
-
Molecular Determinants of Chronic Venous Disease: A Comprehensive Review.Int J Mol Sci. 2023 Jan 18;24(3):1928. doi: 10.3390/ijms24031928. Int J Mol Sci. 2023. PMID: 36768250 Free PMC article. Review.
References
-
- Raetz J., Wilson M., Collins K. Varicose veins: diagnosis and treatment. American Family Physician . 2019;99(11):682–688. - PubMed
-
- Nüllen H., Noppeney T. Diagnosis and treatment of varicose veins. Part 1: definition, epidemiology, etiology, classification, clinical aspects, diagnostic and indications. Der Chirurg; Zeitschrift fur Alle Gebiete der Operativen Medizen . 2010;81:1035–1044. doi: 10.1007/s00104-009-1865-y. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

