RadicalSAM.org: A Resource to Interpret Sequence-Function Space and Discover New Radical SAM Enzyme Chemistry
- PMID: 36119373
- PMCID: PMC9477430
- DOI: 10.1021/acsbiomedchemau.1c00048
RadicalSAM.org: A Resource to Interpret Sequence-Function Space and Discover New Radical SAM Enzyme Chemistry
Abstract
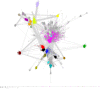
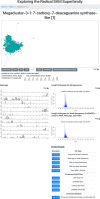
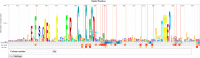
The radical SAM superfamily (RSS), arguably the most functionally diverse enzyme superfamily, is also one of the largest with ~700K members currently in the UniProt database. The vast majority of the members have uncharacterized enzymatic activities and metabolic functions. In this Perspective, we describe RadicalSAM.org, a new web-based resource that enables a user-friendly genomic enzymology strategy to explore sequence-function space in the RSS. The resource attempts to enable identification of isofunctional groups of radical SAM enzymes using sequence similarity networks (SSNs) and the genome context of the bacterial, archaeal, and fungal members provided by genome neighborhood diagrams (GNDs). Enzymatic activities and in vivo functions frequently can be inferred from genome context given the tendency for genes of related function to be clustered. We invite the scientific community to use RadicalSAM.org to (i) guide their experimental studies to discover new enzymatic activities and metabolic functions, (ii) contribute experimentally verified annotations to RadicalSAM.org to enhance the ability to predict novel activities and functions, and (iii) provide suggestions for improving this resource.
Keywords: Radical SAM superfamily; functional assignment; genome neighborhood diagrams; genomic enzymology; isofunctional families; sequence similarity networks; web resource.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



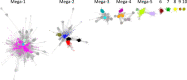








References
-
- Sofia H. J.; Chen G.; Hetzler B. G.; Reyes-Spindola J. F.; Miller N. E. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001, 29, 1097–1106. 10.1093/nar/29.5.1097. - DOI - PMC - PubMed
-
- Harrison P. W.; Ahamed A.; Aslam R.; Alako B. T. F.; Burgin J.; Buso N.; Courtot M.; Fan J.; Gupta D.; Haseeb M.; Holt S.; Ibrahim T.; Ivanov E.; Jayathilaka S.; Balavenkataraman Kadhirvelu V.; Kumar M.; Lopez R.; Kay S.; Leinonen R.; Liu X.; O’Cathail C.; Pakseresht A.; Park Y.; Pesant S.; Rahman N.; Rajan J.; Sokolov A.; Vijayaraja S.; Waheed Z.; Zyoud A.; Burdett T.; Cochrane G. The European Nucleotide Archive in 2020. Nucleic Acids Res. 2021, 49, D82–D85. 10.1093/nar/gkaa1028. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
