Comprehensive analysis of TP53 and SPOP mutations and their impact on survival in metastatic prostate cancer
- PMID: 36119488
- PMCID: PMC9471084
- DOI: 10.3389/fonc.2022.957404
Comprehensive analysis of TP53 and SPOP mutations and their impact on survival in metastatic prostate cancer
Abstract
Background: Although TP53 and SPOP are frequently mutated in metastatic prostate cancer (PCa), their prognostic value is ambiguous, and large sample studies are lacking, especially when they co-occur with other genetic alterations.
Methods: Genomic data and patients' clinical characteristics in PCa were downloaded from the cBioPortal database. We extensively analyzed other gene alterations in different mutation status of TP53 and SPOP. We further subdivided TP53 and SPOP mutation into subgroups based on different mutation status, and then evaluated the prognostic value. Two classification systems for TP53 survival analysis were used.
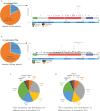
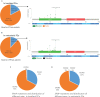
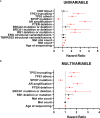
Results: A total of 2,172 patients with PCa were analyzed in our study, of which 1,799 were metastatic PCa patients. The mutual exclusivity analysis showed that TP53 and SPOP mutation has a strong mutual exclusion (p<0.001). In multivariable analysis, truncating TP53 mutations (HR=1.773, 95%CI:1.403-2.239, p<0.001) and other TP53 mutations(HR=1.555, 95%CI:1.267-1.908, p<0.001) were independent negative prognostic markers in metastatic PCa, whereas SPOP mutations(HR=0.592, 95%CI:0.427-0.819, p<0.001) were an independent prognostic factor for better prognosis. Mutations in TP53 were significantly associated with wild-type status for SPOP and CDK12, structural variants/fusions for TMPRSS2 and ERG, AR amplification and PTEN deletion (p<0.001). And truncating TP53 mutations have higher AR amplification rates than other TP53 mutations (p=0.022). Consistently, truncating TP53 mutations had a worse prognosis than other TP53 mutations (p<0.05). Then Kaplan-Meier survival curve showed that Co-occurring TP53 mutations in AR amplification or PTEN deletion tumors significantly reduced survival (p<0.05). Furthermore, those with SPOP-mutant tumors with co-occurring TP53 truncating mutations had shorter overall survival than those with SPOP-mutant tumors with wild-type or other TP53 mutations.
Conclusions: This study found that TP53 and SPOP mutations were mutually exclusive and both were independent prognostic markers for metastatic PCa. Genomic alteration and survival analysis revealed that TP53 and SPOP mutations represented distinct molecular subtypes. Our data suggest that molecular stratification on the basis of TP53 and SPOP mutation status should be implemented for metastatic PCa to optimize and modify clinical decision-making.
Keywords: SPOP; TP53; biomarkers; metastatic prostate cancer; mutation; prognosis.
Copyright © 2022 Zhou, Lai, Peng, Tang, Chen, Li, Huang and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Clinical and genomic features of SPOP-mutant prostate cancer.Prostate. 2022 Feb;82(2):260-268. doi: 10.1002/pros.24269. Epub 2021 Nov 15. Prostate. 2022. PMID: 34783071 Free PMC article.
-
Metastatic prostate cancer is associated with distinct higher frequency of genetic mutations at diagnosis.Urol Oncol. 2023 Nov;41(11):455.e7-455.e15. doi: 10.1016/j.urolonc.2023.09.014. Epub 2023 Oct 13. Urol Oncol. 2023. PMID: 37838503
-
Comprehensive Analysis of TP53 and KEAP1 Mutations and Their Impact on Survival in Localized- and Advanced-Stage NSCLC.J Thorac Oncol. 2022 Jan;17(1):76-88. doi: 10.1016/j.jtho.2021.08.764. Epub 2021 Sep 30. J Thorac Oncol. 2022. PMID: 34601169
-
Updates on SPOP Gene Mutations in Prostate Cancer and Computational Insights From TCGA cBioPortal Database.Scientifica (Cairo). 2025 May 20;2025:4084224. doi: 10.1155/sci5/4084224. eCollection 2025. Scientifica (Cairo). 2025. PMID: 40432836 Free PMC article. Review.
-
DNA alterations in the tumor genome and their associations with clinical outcome in prostate cancer.Asian J Androl. 2016 Jul-Aug;18(4):533-42. doi: 10.4103/1008-682X.177120. Asian J Androl. 2016. PMID: 26975494 Free PMC article. Review.
Cited by
-
Exploring The Prognostic Significance of SET-Domain Containing 2 (SETD2) Expression in Advanced and Castrate-Resistant Prostate Cancer.Cancers (Basel). 2024 Apr 8;16(7):1436. doi: 10.3390/cancers16071436. Cancers (Basel). 2024. PMID: 38611113 Free PMC article.
-
Integrated analysis of single-cell and bulk RNA sequencing identifies a signature based on macrophage marker genes involved in prostate cancer prognosis and treatment responsiveness.Funct Integr Genomics. 2023 Apr 3;23(2):115. doi: 10.1007/s10142-023-01037-9. Funct Integr Genomics. 2023. PMID: 37010617
-
Cuproptosis-related lncRNAs emerge as a novel signature for predicting prognosis in prostate carcinoma and functional experimental validation.Front Immunol. 2024 Oct 28;15:1471198. doi: 10.3389/fimmu.2024.1471198. eCollection 2024. Front Immunol. 2024. PMID: 39530098 Free PMC article.
-
TP53 Deficiency in the Natural History of Prostate Cancer.Cancers (Basel). 2025 Feb 14;17(4):645. doi: 10.3390/cancers17040645. Cancers (Basel). 2025. PMID: 40002239 Free PMC article. Review.
-
PTEN Mutations Associated with Increased Recurrence and Decreased Survival in Patients with Prostate Cancer Spinal Metastasis.Curr Oncol. 2025 Jun 4;32(6):331. doi: 10.3390/curroncol32060331. Curr Oncol. 2025. PMID: 40558274 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

