Genomic mapping of copy number variations influencing immune response in breast cancer
- PMID: 36119512
- PMCID: PMC9476651
- DOI: 10.3389/fonc.2022.975437
Genomic mapping of copy number variations influencing immune response in breast cancer
Abstract
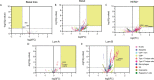
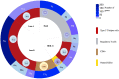
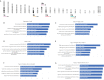
Identification of genomic alterations that influence the immune response within the tumor microenvironment is mandatory in order to identify druggable vulnerabilities. In this article, by interrogating public genomic datasets we describe copy number variations (CNV) present in breast cancer (BC) tumors and corresponding subtypes, associated with different immune populations. We identified regulatory T-cells associated with the Basal-like subtype, and type 2 T-helper cells with HER2 positive and the luminal subtype. Using gene set enrichment analysis (GSEA) for the Type 2 T-helper cells, the most relevant processes included the ERBB2 signaling pathway and the Fibroblast Growth Factor Receptor (FGFR) signaling pathway, and for CD8+ T-cells, cellular response to growth hormone stimulus or the JAK-STAT signaling pathway. Amplification of ERBB2, GRB2, GRB7, and FGF receptor genes strongly correlated with the presence of type 2 T helper cells. Finally, only 8 genes were highly upregulated and present in the cellular membrane: MILR1, ACE, DCSTAMP, SLAMF8, CD160, IL2RA, ICAM2, and SLAMF6. In summary, we described immune populations associated with genomic alterations with different BC subtypes. We observed a clear presence of inhibitory cells, like Tregs or Th2 when specific chromosomic regions were amplified in basal-like or HER2 and luminal groups. Our data support further evaluation of specific therapeutic strategies in specific BC subtypes, like those targeting Tregs in the basal-like subtype.
Keywords: CNVs; Gene Amplification; breast cancer; immune response; new surface targets.
Copyright © 2022 López-Cade, García-Barberán, Cabañas Morafraile, Díaz-Tejeiro, Saiz-Ladera, Sanvicente, Pérez Segura, Pandiella, Győrffy and Ocaña.
Conflict of interest statement
AO is a consultant of Servier and a former employee of Symphogen. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Landscape of somatic allelic imbalances and copy number alterations in HER2-amplified breast cancer.Breast Cancer Res. 2011;13(6):R129. doi: 10.1186/bcr3075. Epub 2011 Dec 14. Breast Cancer Res. 2011. PMID: 22169037 Free PMC article.
-
Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery.PLoS One. 2009 Jul 3;4(7):e6146. doi: 10.1371/journal.pone.0006146. PLoS One. 2009. PMID: 19582160 Free PMC article.
-
Comprehensive Analysis of Regulatory Factors and Immune-Associated Patterns to Decipher Common and BRCA1/2 Mutation-Type-Specific Critical Regulation in Breast Cancer.Front Cell Dev Biol. 2021 Oct 18;9:750897. doi: 10.3389/fcell.2021.750897. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34733851 Free PMC article.
-
Ki67 and lymphocytes in the pretherapeutic core biopsy of primary invasive breast cancer: positive markers of therapy response prediction and superior survival.Horm Mol Biol Clin Investig. 2017 Sep 22;32(2):/j/hmbci.2017.32.issue-2/hmbci-2017-0022/hmbci-2017-0022.xml. doi: 10.1515/hmbci-2017-0022. Horm Mol Biol Clin Investig. 2017. PMID: 28937963 Review.
-
Is There a Benefit of HER2-Positive Breast Cancer Subtype Determination in Clinical Practice?Klin Onkol. 2019 Winter;32(1):25-30. doi: 10.14735/amko2019. Klin Onkol. 2019. PMID: 30764626 Review. English.
Cited by
-
GRB7 Plays a Vital Role in Promoting the Progression and Mediating Immune Evasion of Ovarian Cancer.Pharmaceuticals (Basel). 2024 Aug 7;17(8):1043. doi: 10.3390/ph17081043. Pharmaceuticals (Basel). 2024. PMID: 39204147 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

