Genome-wide identification and multiple abiotic stress transcript profiling of potassium transport gene homologs in Sorghum bicolor
- PMID: 36119582
- PMCID: PMC9478208
- DOI: 10.3389/fpls.2022.965530
Genome-wide identification and multiple abiotic stress transcript profiling of potassium transport gene homologs in Sorghum bicolor
Abstract
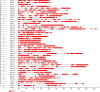
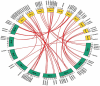
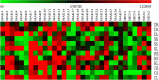
Potassium (K+) is the most abundant cation that plays a crucial role in various cellular processes in plants. Plants have developed an efficient mechanism for the acquisition of K+ when grown in K+ deficient or saline soils. A total of 47 K+ transport gene homologs (27 HAKs, 4 HKTs, 2 KEAs, 9 AKTs, 2 KATs, 2 TPCs, and 1 VDPC) have been identified in Sorghum bicolor. Of 47 homologs, 33 were identified as K+ transporters and the remaining 14 as K+ channels. Chromosome 2 has been found as the hotspot of K+ transporters with 9 genes. Phylogenetic analysis revealed the conservation of sorghum K+ transport genes akin to Oryza sativa. Analysis of regulatory elements indicates the key roles that K+ transport genes play under different biotic and abiotic stress conditions. Digital expression data of different developmental stages disclosed that expressions were higher in milk, flowering, and tillering stages. Expression levels of the genes SbHAK27 and SbKEA2 were higher during milk, SbHAK17, SbHAK11, SbHAK18, and SbHAK7 during flowering, SbHAK18, SbHAK10, and 23 other gene expressions were elevated during tillering inferring the important role that K+ transport genes play during plant growth and development. Differential transcript expression was observed in different tissues like root, stem, and leaf under abiotic stresses such as salt, drought, heat, and cold stresses. Collectively, the in-depth genome-wide analysis and differential transcript profiling of K+ transport genes elucidate their role in ion homeostasis and stress tolerance mechanisms.
Keywords: HAK/KT/KUP; K+ channels; K+ transporters; KEA; Sorghum; Trk/HKT.
Copyright © 2022 Anil Kumar, Hima Kumari, Nagaraju, Sudhakar Reddy, Durga Dheeraj, Mack, Katam and Kavi Kishor.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Aksu G., Altay H. (2020). The effects of potassium applications on drought stress in sugar beet. Sugar Tech 22 1092–1102. 10.26900/jsp.4.013 - DOI
-
- Amrutha R. N., Sekhar P. N., Varshney R. K., Kavi Kishor P. B. (2007). Genome-wide analysis and identification of genes related to potassium transporter families in rice (Oryza sativa L.). Plant Sci. 172 708–721. 10.1016/j.plantsci.2006.11.019 - DOI
LinkOut - more resources
Full Text Sources

