Bacterial genome reductions: Tools, applications, and challenges
- PMID: 36120530
- PMCID: PMC9473318
- DOI: 10.3389/fgeed.2022.957289
Bacterial genome reductions: Tools, applications, and challenges
Abstract
Bacterial cells are widely used to produce value-added products due to their versatility, ease of manipulation, and the abundance of genome engineering tools. However, the efficiency of producing these desired biomolecules is often hindered by the cells' own metabolism, genetic instability, and the toxicity of the product. To overcome these challenges, genome reductions have been performed, making strains with the potential of serving as chassis for downstream applications. Here we review the current technologies that enable the design and construction of such reduced-genome bacteria as well as the challenges that limit their assembly and applicability. While genomic reductions have shown improvement of many cellular characteristics, a major challenge still exists in constructing these cells efficiently and rapidly. Computational tools have been created in attempts at minimizing the time needed to design these organisms, but gaps still exist in modelling these reductions in silico. Genomic reductions are a promising avenue for improving the production of value-added products, constructing chassis cells, and for uncovering cellular function but are currently limited by their time-consuming construction methods. With improvements to and the creation of novel genome editing tools and in silico models, these approaches could be combined to expedite this process and create more streamlined and efficient cell factories.
Keywords: bacteria; genome engineering; genome reduction; minimal genome; synthetic biology.
Copyright © 2022 LeBlanc and Charles.
Conflict of interest statement
TCC is a major shareholder of Metagenom Bio Life Science Inc. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
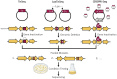
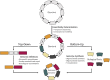

References
Publication types
LinkOut - more resources
Full Text Sources

