A non-canonical scaffold-type E3 ligase complex mediates protein UFMylation
- PMID: 36121123
- PMCID: PMC9627666
- DOI: 10.15252/embj.2022111015
A non-canonical scaffold-type E3 ligase complex mediates protein UFMylation
Abstract
Protein UFMylation, i.e., post-translational modification with ubiquitin-fold modifier 1 (UFM1), is essential for cellular and endoplasmic reticulum homeostasis. Despite its biological importance, we have a poor understanding of how UFM1 is conjugated onto substrates. Here, we use a rebuilding approach to define the minimal requirements of protein UFMylation. We find that the reported cognate E3 ligase UFL1 is inactive on its own and instead requires the adaptor protein UFBP1 to form an active E3 ligase complex. Structure predictions suggest the UFL1/UFBP1 complex to be made up of winged helix (WH) domain repeats. We show that UFL1/UFBP1 utilizes a scaffold-type E3 ligase mechanism that activates the UFM1-conjugating E2 enzyme, UFC1, for aminolysis. Further, we characterize a second adaptor protein CDK5RAP3 that binds to and forms an integral part of the ligase complex. Unexpectedly, we find that CDK5RAP3 inhibits UFL1/UFBP1 ligase activity in vitro. Results from reconstituting ribosome UFMylation suggest that CDK5RAP3 functions as a substrate adaptor that directs UFMylation to the ribosomal protein RPL26. In summary, our reconstitution approach reveals the biochemical basis of UFMylation and regulatory principles of this atypical E3 ligase complex.
Keywords: E3 ligase; enzyme substrate; post-translational modification; ribosome; ubiquitin-like modifier.
© 2022 The Authors. Published under the terms of the CC BY 4.0 license.
Figures

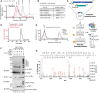
- A
Comparison of size exclusion chromatography profiles of UFL1 expressed alone (Red) and co‐expressed UFL1/UFBP1 complex (Black) run under identical buffer conditions on a Superdex™ 200 Increase 10/300 GL column. Molecular weight standards are shown in gray. Fractions corresponding to each peak were collected and analyzed on 4–12% denaturing SDS–PAGE followed by Coomassie staining, which is shown below.
- B
Mass photometry analysis of co‐purified UFL1/UFBP1 complex. The theoretical and experimental molecular weights are indicated above.
- C
Immunoblot comparing UFL1 autoUFMylation (Top) and UFM1 chain synthesis (Middle) in the presence of UFL1 expressed alone and in complex with UFBP1. (Bottom) Assays to monitor UFMylation of substrates using purified TRIP4. Reaction products were separated on a 4–12% SDS–PAGE gel under reducing conditions and analyzed by immunoblotting using indicated antibodies.
- D
In vitro reconstitution of 60S ribosome UFMylation. Purified 60S ribosome was incubated with either UFL1 on its own or UFL1/UFBP1 complex together with UBA5, UFC1, and UFM1 for indicated time points at 30°C.
- E
Immunoblot showing the formation of free UFM1 chains in the presence (lane 3 and 4) and absence of UFL1/UFBP1 (lane 1 and 2). (Bottom) Graphical representation of the linkage composition of di‐UFM1 chains obtained from LC–MS/MS analysis.
- F
Immunoblot analyzing polyUFMylation (top) and UFL1 autoUFMylation (bottom) in the presence of indicated Lys to Arg (K‐R) and Lys‐less (K0) mutants of UFM1.
- G
Immunoblot showing in vitro UFMylation assay to check for the formation of di‐UFM1 chains in the presence of WT, Lys‐less (K0), and single lys mutants (K only) of UFM1.
- A
Size exclusion chromatography profile of UFL1 run on Superdex™ 200 Increase 10/300 GL column (shown in red). Overlay of UV chromatogram of molecular weight standards of different sizes (shown in grey) run under same buffer conditions.
- B
Results obtained from ULTIMATE Y2H™ screening (Hybrigenics) for binary interactions with UFL1. Asterisk (*) denotes proteins hits obtained with low confidence.
- C
Schematic describing the strategy for co‐expression and purification of UFL1/UFBP1 complex.
- D
SEC‐MALS analysis of UFL1/UFBP1 complex. The theoretical and observed molecular weights are indicated above.
- E
Analytical gel filtration chromatography analysis showing UV traces of UFL1 and UFBP1 run on their own and a mixture containing UFL1 and UFBP1. Superdex™ 200 Increase 3.2/300 column was used for analysis.
- F
UFBP1 does not activate UFL1 when added exogenously. In vitro UFMylation assays to compare the E3 ligase activity of UFL1 and UFBP1 expressed alone and together as a complex. 0.25 μM UBA5, 5 μM UFC1 and 10 μM UFM1 was incubated with 1 μM UFL1 or 1 μM UFBP1 or 1 μM UFL1/UFBP1 complex for 1 h at 37°C in the buffer containing 50 mM HEPES 7.5, 0.5 mM DTT, 10 mM MgCl2 and 10 mM ATP. The reaction was stopped by addition of 3× SDS loading dye and run on a 4–12% denaturing SDS PAGE gel under reducing conditions and immunoblotting was performed using indicated antibodies.
- G
MS2 spectra showing the peptide derived from in vitro UFMylation assay showing the VG‐remnant on K69 of UFM1.
- A
Schematic showing the workflow for substrate‐independent single turnover lysine discharge assays.
- B
Coomassie‐stained gels monitoring the discharge of UFM1 from UFC1 in the presence of indicated free amino acids. UBE2D3 is used as a positive control for lysine and cysteine discharge.
- C
Multiple sequence alignment of 1–410 a.a. region of UFL1 from various organisms to highlight the degree of conservation of cysteine residues in this region.
- D
Immunoblot showing in vitro UFMylation assays in the presence of Cys to Ala (indicated as C‐A) mutants of UFL1 to check for UFBP1 UFMylation (top), UFL1 autoUFMylation (middle), and substrate UFMylation (bottom).
- E
Single turnover lysine discharge assays in the presence and absence of full‐length UFL1/UFBP1 complex. The reaction was stopped by the addition of nonreducing SDS loading buffer (1× final) and analyzed for discharge on a 4–12% SDS–PAGE gel under nonreducing conditions. Top gel—LI‐COR scan of fluorescently labeled UFM1 (UFM1*); bottom gel—Coomassie stained.

- A
Single turnover assay to monitor discharge of Ubiquitin from UBE2D3 and UBE2L3 in the presence of free amino acids. “0” indicates time 0.
- B
Single turnover lysine discharge assays using UBE2D3 and UFC1 in the presence of increasing concentration of free Lysine.
- C
Time‐dependent analysis of discharge of UFM1 from UFC1 in the presence of high concentration of free lysine (150 mM).
- D, E
Time‐dependent analysis of discharge of Ubiquitin from UBE2D3 and UBE2L3 in the presence of 150 mM Lysine.
- F
Composite Foldindex profile of UFL1 showing folding propensity of different regions of UFL1 to aid in construct design for soluble protein expression.
- G
Size exclusion chromatography profile of MBP‐UFL11‐410 (dark blue) and His6‐UFL11‐410/UFBP129‐end (orange) run on Superdex™ 200 Increase 3.2/300 column. Approximately 20 μg of sample was used for analysis. (Right) Coomassie stained gel showing the purity of the proteins.
- H
In vitro UFMylation assay to check for E3 ligase activity of UFL11‐410/UFBP129‐end. Full length UFL1/UFBP1 complex is used as a positive control.
- I
Time dependent analysis of transthiolation activity of UFL1. Reaction products were analysed on a 4–12% SDS PAGE gel under reducing (lane 1–5) and non‐reducing conditions (lanes 6–10).
- J
In vitro UFMylation assays to check for formation of di‐UFM1 chains by minimal reconstitution using UBA5, UFC1 and UFM1 in the presence of MgCl2 and ATP.
- K
Immunoblot to check for the presence of free UFM1 chains with and without treatment of UFSP2. In vitro UFMylation products generated as shown in (E) was incubated with UFSP2 (2 μM) for 1 h at 37°C. The reaction was stopped by addition of SDS‐loading buffer (1× final) and run on a 4–12% SDS PAGE gel under reducing conditions followed by immunoblotting using indicated antibodies to check for the disappearance of polyUFMylated products especially di‐UFM1.
- L
Graphical representation showing the composition of linkage forms of di‐UFM1 chains formed by minimal reconstitution of UBA5 and UFC1.

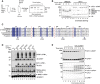
- A
Cartoon representation of predicted full‐length human UFL1/UFBP1 complex using AlphaFfold. UFL1 and UFBP1 are shown in blue and teal colors, respectively. The dimeric interface of UFL1 and UFBP1 is highlighted in the dotted box. WH, Winged helix; pWH, Partial Winged helix; NTR, N‐terminal region; CTR, C‐terminal region.
- B
View of the dimeric interface of UFL1/UFBP1 complex highlighting the formation of a full composite WH domain by two partial winged helix domains of UFL1 (blue) and UFBP1 (teal).
- C
Close‐up view of the interface of UFL1/UFBP1 complex highlighting the interactions at the pWH‐pWH′ domain interface centered on UFL1 Leu45 (L45) and UFBP1 Ile283 (I283).
- D
Immunoblot from immunoprecipitation (IP) assays to validate the predicted model of UFL1/UFBP1 interaction in cells. UFL1WT‐3xFLAG and UFL1L45R‐3xFLAG were transiently overexpressed along with UFBP1‐SBP in HEK293T‐UFL1 KO cells and subjected to separate pulldowns using anti‐FLAG antibody or streptavidin. Immunoprecipitated material was run on a 4–12% SDS–PAGE gel and analyzed by immunoblotting using indicated antibodies.
- E
Schematic representation of UFL1/UFBP1 constructs with different domain boundaries designed to identify the minimal catalytic region of the UFL1/UFBP1 complex required for aminolysis.
- F
Quantitative representation of single turnover lysine discharge assays to identify the minimal boundaries of UFL1 (left) and UFBP1 (right) required for activation of UFC1 (n = 3, mean ± SD). A representative gel image used for the quantitative analysis is shown in Fig EV4A and B.
- G
Immunoblot showing in vitro UFMylation assay to monitor the formation of free di‐UFM1 chains in the presence of UFL1/UFBP1 complexes bearing different domain boundaries.
- H
Immunoblot of in vitro assay monitoring UFMylation of 60S Ribosome in the presence of different UFL1/UFBP1 truncations as depicted in (E).

- A
Predicted Aligned Error (PAE) scores of UFL1/UFBP1 models predicted using AlphaFold.
- B
List of structurally similar proteins from the PDB25 database predicted using UFL1 WH1 (52–115 a.a) domain as query structure using DALI server.
- C
Overlaid structures of Winged helix 1 (WH1) domain of UFL1 (52–115 a.a) and top three hits obtained from DALI search shown in cartoon representation.
- D
Comparison of winged helix domains of UFL1 (shown in blue) and UFBP1 (shown in teal) to highlight their structural similarities.
- E
SEC elution profiles showing that full length UFL1/UFBP1 preferentialy interacts with charged E2. UFL1/UFBP1 complex was incubated with UFC1‐O‐UFM1 at a 1:1 molar ratio for 20 min at 4°C and loaded on a Superdex™ 200 Increase 3.2/300 column. The fractions corresponding to each peak were collected and separated on a 4–12% SDS PAGE gel followed by Coomassie staining.
- F
Minimal catalytic region is sufficient for interaction with charged UFC1. UFL1(1‐179)/UFBP1(1‐116) complex was incubated with UFC1‐O‐UFM1 at the molar ratio of 1:1 for 20 min at 4°C and analysed by analytical size exclusion chromatography as described in (E).
- G
Coomassie stained gel showing purity of different UFL1/UFBP1 truncations.

- A
Lysine discharge assays to check for activation of UFC1 in the presence of different UFL1 truncations. Top gel: LI‐COR scan of fluorescently labelled UFM1 (UFM1*); bottom gel—Coomassie stained (representative of three independent experiments); Related to Fig 3F.
- B
Lysine discharge assays as in (A) in the absence of NTR of UFBP1. Top gel: LI‐COR scan of fluorescently labelled UFM1 (UFM1*); bottom gel—Coomassie stained (representative of three independent experiments); Related to Fig 3F.
- C
Coomassie stained gel showing analysis of in vitro UFMylation reaction products to check for the formation of stable oxy‐ester linked UFC1‐UFM1 conjugate (UFC1‐O‐UFM1).
- D
Chromatogram obtained from SEC analysis using HiLoad™ 16/60 Superdex™ 75 pg column. (Bottom left) The fractions collected were run on an SDS‐PAGE gel to identify fractions that contained pure UFC1‐O‐UFM1. (Bottom right) Coomassie stained gel showing analysis of purified UFC1‐O‐UFM1 product to check for homogeneity.
- E
Quality check to analyse if UFC1‐UFM1 conjugate is linked through an oxy‐ester linkage by alkaline hydrolysis.
- F, G
Substrate UFMylation assays to check for UFMylation of MRE11 and Histone H4 respectively in the presence of different UFL1/UFBP1 truncations.
- H
Role of UFBP1 in substrate UFMylation. (Top) Comparison of E3 ligase activity of UFL1/UFBP129‐end and UFL1/UFBP1207‐end using Histone H4. (Bottom) Comparison of E3 ligase activity of UFL11‐179/UFBP129‐end and UFL11‐179/UFBP1207‐end using purified Histone H4.
- I
Schematic showing the tandem WH domains which constitute the minimal ligase domain.
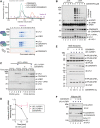
- A
CDK5RAP3 forms a stable complex with UFL1/UFBP1 in vitro. Thirty microliter of UFL1/UFBP1 was mixed with 15 μg of CDK5RAP3 and loaded on a Superdex™ 200 Increase 3.2/300 column analytical gel filtration column. (Bottom) Fractions were collected and analyzed on 4–12% SDS–PAGE under reducing conditions and visualized by Coomassie staining.
- B
In vitro UFMylation assay in the presence of increasing concentrations of CDK5RAP3 to monitor the E3 ligase activity of UFL1/UFBP1 complex. The reaction products were run on a 4–12% SDS–PAGE gel under reducing conditions and immunoblotting was performed using indicated antibodies.
- C
Single turnover lysine discharge assays to check for activation of UFC1 by UFL1/UFBP1 in the presence and absence of CDK5RAP3. The reaction products were run on a 4–12% SDS–PAGE and visualized by Oriole staining.
- D
Quantification of the discharge of UFM1 from UFC1 in the presence and absence of CDK5RAP3 as seen in (D), n = 3 biological replicates, mean ± SD.
- E
Substrate UFMylation assays using purified 60S Ribosomes in the presence of an increasing concentration of CDK5RAP3. Reaction was performed for 10 min, stopped by the addition of SDS Loading dye, and analyzed on an SDS–PAGE gel under reducing conditions followed by immunoblotting with indicated antibodies.
- F
Immunoblot analysis monitoring UFMylation of Histone H4 in the presence of increasing concentrations of CDK5RAP3 (1, 2, 3 μM). UFL1/UFBP1 concentrations—1 μM.

- A
Mass photometry analysis showing the experimental molecular weight of UFL1/UFBP1/CDK5RAP3 complex.
- B
In vitro UFMylation assay to compare the E3 ligase activity of UFL1/UFBP1 mixed with CDK5RAP3 and preassembled ternary E3 ligase complex containing UFL1/UFBP1/CDK5RAP3. Ponceau‐stained nitrocellulose membrane is shown below to indicate the amounts of reaction components used in the assay.
- C
UV traces of gel filtration chromatogram showing co‐migration of (UFL1/UFBP1)/CDK5RAP3/UFC1‐O‐UFM1 complex. Approximately, 200 μl of sample containing (UFL1/UFBP1)/CDK5RAP3/UFC1‐O‐UFM1 in the molar ratio of 1:1.5:3 was mixed and incubated at 4°C for 1 h and loaded onto a Superdex™ 200 Increase 10/300 GL column. The fractions were collected and analysed on a 4–12% SDS PAGE and visualized by Coomassie staining.
- D
Pulldown assay to check for interaction of UFL1/UFBP1 with charged UFC1 in the presence of absence of CDK5RAP3. Around 10 μM of Untagged UFC1 and 10 μM of UFC1‐O‐UFM1 were mixed with 5 μM of UFL1/UFBP1 complex in the presence and absence of CDK5RAP3.
- E
Single turnover lysine discharge assays to check for UFC1 discharge in the presence of UFL1/UFBP1 mixed with CDK5RAP3 and preassembled UFL1/UFBP1/CDK5RAP3 complex. The reaction products were run on a 4–12% SDS PAGE analysis and visualized by Coomassie staining.
- F
In vitro UFMylation assay to monitor UFMylation of purified substrates namely MRE11A (left) and TRIP4 (right) in the presence of increasing concentration of CDK5RAP3 (1, 2, 3 μM). UFL1/UFBP1 concentration—1 μM.

- A
Comparison of catalytic sites of UBE2D3 (PDB ID:5EGG, shown in green) and UFC1 (PDB ID:2Z6O, shown in pink) shown as cartoons. The residues of the conserved HPN and TAK motif of UBE2D3 and UFC1, respectively, are shown in ball and stick representation.
- B
Immunoblot showing the in vitro UFMylation assay to compare the activity of UFC1 Wild type and mutants in the presence of UFL1/UFBP1 complex.
- C
In vitro UFMylation assay to compare the activity of UFC1 Wild type and UFC1 T106I in the presence of UFL1/UFBP1 complex.
- D
Quantification of lysine and cysteine discharge of UFM1 from UFC1 WT and other indicated mutants (n = 3 biological replicates, mean ± SD). The first time point in cysteine discharge assays indicated as “0” represents reaction products before the addition of cysteine. Following the addition of 5 mM cysteine, the reaction was incubated at RT for the specified time duration. The reaction was stopped by the addition of nonreducing SDS loading buffer (1× final) and analyzed as described above. A representative gel used for lysine quantification is given in Appendix Fig S1C and D.
- E
Substrate UFMylation assays using purified 60S Ribosomes in the presence of UFC1 Wild type and different mutants of UFC1.
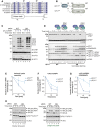
- A
Multiple sequence alignment of UFC1 homologs from various organisms. A graphical representation of the secondary structure is extracted from the crystal structure of UFC1 (PDB ID:2Z6O).
- B
Schematic representation of domain features of UFC1WT and UFC1ΔN.
- C
In vitro UFMylation assays in the presence of Wild type UFC1 (UFC1WT) and UFC1 lacking the N‐terminal helix (UFC1ΔN).
- D
Assay to compare Lys discharge activities of UFC1WT and UFC1ΔN on its own, in the presence of UFL1/UFBP1 or in the presence of preassembled UFL1/UFBP1/CDK5RAP3 complex. Top gel: LI‐COR scan of fluorescently labeled UFM1 (UFM1*); bottom gel—Coomassie stained (Representative of three independent experiments).
- E–G
(E) Quantitative analysis of the intrinsic Lys reactivity (Lysine 25 mM) of UFC1WT and in UFC1ΔN in the absence of E3 ligase; (F) in the presence of UFL1/UFBP1; (G) in the presence of preassembled UFL1/UFBP1/CDK5RAP3 (E–G: n = 3 biological replicates, mean ± SD).
- H
Effect of deletion of N‐term helix of UFC1 in 60S ribosome UFMylation. Purified 60S ribosomes (50 nM) were incubated with 0.5 μM UBA5, 1 μM UFM1, 100 nM UFL1/UFBP1 complex in the presence of either 1 μM UFC1WT or 1 μM UFC1ΔN at 30°C. The reaction was stopped at indicated time points and run on a 4–12% SDS–PAGE gel under reducing conditions followed by immunoblotting using indicated antibodies (Right—shorter time duration. Left—Longer time duration).
References
-
- Aravind L, Anantharaman V, Balaji S, Babu M, Iyer L (2005) The many faces of the helix‐turn‐helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29: 231–262 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

