African Ancestry-Associated Gene Expression Profiles in Triple-Negative Breast Cancer Underlie Altered Tumor Biology and Clinical Outcome in Women of African Descent
- PMID: 36121736
- PMCID: PMC9627137
- DOI: 10.1158/2159-8290.CD-22-0138
African Ancestry-Associated Gene Expression Profiles in Triple-Negative Breast Cancer Underlie Altered Tumor Biology and Clinical Outcome in Women of African Descent
Abstract
Women of sub-Saharan African descent have disproportionately higher incidence of triple-negative breast cancer (TNBC) and TNBC-specific mortality across all populations. Population studies show racial differences in TNBC biology, including higher prevalence of basal-like and quadruple-negative subtypes in African Americans (AA). However, previous investigations relied on self-reported race (SRR) of primarily U.S. populations. Due to heterogeneous genetic admixture and biological consequences of social determinants, the true association of African ancestry with TNBC biology is unclear. To address this, we conducted RNA sequencing on an international cohort of AAs, as well as West and East Africans with TNBC. Using comprehensive genetic ancestry estimation in this African-enriched cohort, we found expression of 613 genes associated with African ancestry and 2,000+ associated with regional African ancestry. A subset of African-associated genes also showed differences in normal breast tissue. Pathway enrichment and deconvolution of tumor cellular composition revealed that tumor-associated immunologic profiles are distinct in patients of African descent.
Significance: Our comprehensive ancestry quantification process revealed that ancestry-associated gene expression profiles in TNBC include population-level distinctions in immunologic landscapes. These differences may explain some differences in race-group clinical outcomes. This study shows the first definitive link between African ancestry and the TNBC immunologic landscape, from an African-enriched international multiethnic cohort. See related commentary by Hamilton et al., p. 2496. This article is highlighted in the In This Issue feature, p. 2483.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures
![Figure 1. Estimated genetic ancestry distribution in an African-enriched TNBC RNA-seq cohort. Genetic ancestry was estimated from genotypes of the ancestry-informed markers obtained from our RNA-seq alignments, in which we have superpopulation ancestry estimations, relative to the 1000 Genomes superpopulation populations (A), and subpopulation ancestry estimations for each individual in our cohort (B). In both A and B, each column represents an individual in the cohort, in which estimated ancestry from a given superpopulation or subpopulation is shown on the y-axis, and the x-axis is annotated by SRR and location. Superpopulation populations in A are East Asian (EAS, red), South Asian (SAS, blue), European (EUR, green), American (AMR, purple), and African (AFR, orange). Subpopulations in B are shown in variations of their corresponding superpopulation population color (i.e., AFR populations are in varying shades of orange), and population codes are reported in Supplementary Table S1. Samples are ordered by decreasing AFR ancestry [x-axis left to right: African/Ghanaian (Ghana), AA (Alabama, Detroit, New York), African/Ethiopian (Ethiopia), EA (Alabama, Detroit, New York), other/declined (New York), and Asian (New York)]. C, Constellation plot showing phylogeny of samples based on ancestry estimations. SRR of samples are indicated by the colored dots (Ghanaian = light blue, AA = light green, Ethiopian = dark blue, EA = dark green, Asian = light pink, and other/declined = dark pink). Site location of samples is annotated next to the colored dots (A = Alabama, USA; D = Detroit, MI, USA; E = Ethiopia; G = Ghana; and N = New York, NY, USA). D, Scatter plot showing inverse correlation of AFR and EUR ancestry in our gene expression cohort.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8baa/9627137/f904d8532c45/2530fig1.gif)
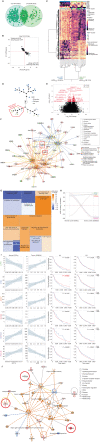



![Figure 6. SRR-unique gene signature enriched in comorbid canonical pathways. A, Venn diagram depicting the overlap of AFR-, EUR-, and SRR-associated genes. DEG, differentially expressed gene. B, Unsupervised hierarchical clustering of the 1,071 SRR-associated genes. AMR, American. C, Unsupervised clustering of 751 genes unique to SRR. In both B and C, columns represent individuals, where SRR and ancestry are shown in the color map at the top, and rows represent genes. Node structure of individuals is shown at the bottom of the heat maps, where clustering was the individual node structure significantly changed between B and C. D, Comparing gene expression values from the node structure in C, we determined enrichment of genes in known canonical pathways that would be associated with environmental exposures and/or potential patient comorbidities. Z-scores indicated predicted activation (positive z-score, orange) or inhibition (negative z-score, blue) of the pathway based on the expression of the genes in the pathway in the directionality of AAs. Black striped bars indicated pathways where no z-score/predication was indicated due to insufficient evidence in the Ingenuity Pathway Analysis knowledge base. The red line indicates a P value cutoff of 0.05 [−log(0.05) = ∼1.3].](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8baa/9627137/bea43a032317/2530fig6.gif)
Comment in
-
Race and Ancestry in Immune Response to Breast Cancer.Cancer Discov. 2022 Nov 2;12(11):2496-2497. doi: 10.1158/2159-8290.CD-22-0852. Cancer Discov. 2022. PMID: 36321309 Free PMC article.
References
-
- Global Burden of Disease Cancer C, Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T, Alizadeh-Navaei R, et al. . Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol 2018;4:1553–68. - PMC - PubMed
-
- Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global cancer in women: burden and trends. Cancer Epidemiol Biomarkers Prev 2017;26:444–57. - PubMed
-
- Newman LA, Kaljee LM. Health disparities and triple-negative breast cancer in African American women: a review. JAMA Surg 2017;152:485–93. - PubMed
-
- Jiagge E, Jibril AS, Chitale D, Bensenhaver JM, Awuah B, Hoenerhoff M, et al. . Comparative analysis of breast cancer phenotypes in African American, White American, and West Versus East African patients: correlation between African ancestry and triple-negative breast cancer. Ann Surg Oncol 2016;23:3843–9. - PubMed
Publication types
MeSH terms
Grants and funding
- L60 MD002419/MD/NIMHD NIH HHS/United States
- U54 CA233465/CA/NCI NIH HHS/United States
- 5U54CA118948/National Cancer Institute (NCI)
- U54-MD007585-26/National Institute on Minority Health and Health Disparities (NIMHD)
- P30 CA013148/CA/NCI NIH HHS/United States
- U54 MD007585/MD/NIMHD NIH HHS/United States
- SC1 GM136521/GM/NIGMS NIH HHS/United States
- R01 CA259396/CA/NCI NIH HHS/United States
- U54 CA118623/CA/NCI NIH HHS/United States
- U54 CA118948/CA/NCI NIH HHS/United States
- P30 CA014089/CA/NCI NIH HHS/United States
- R01 CA259396-01/National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

