Precision genome editing in the eye
- PMID: 36122230
- PMCID: PMC9522375
- DOI: 10.1073/pnas.2210104119
Precision genome editing in the eye
Abstract
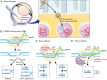
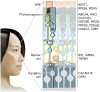
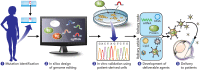
CRISPR-Cas-based genome editing technologies could, in principle, be used to treat a wide variety of inherited diseases, including genetic disorders of vision. Programmable CRISPR-Cas nucleases are effective tools for gene disruption, but they are poorly suited for precisely correcting pathogenic mutations in most therapeutic settings. Recently developed precision genome editing agents, including base editors and prime editors, have enabled precise gene correction and disease rescue in multiple preclinical models of genetic disorders. Additionally, new delivery technologies that transiently deliver precision genome editing agents in vivo offer minimized off-target editing and improved safety profiles. These improvements to precision genome editing and delivery technologies are expected to revolutionize the treatment of genetic disorders of vision and other diseases. In this Perspective, we describe current preclinical and clinical genome editing approaches for treating inherited retinal degenerative diseases, and we discuss important considerations that should be addressed as these approaches are translated into clinical practice.
Keywords: eye; genome editing; retina; retinal degeneration.
Conflict of interest statement
Competing interest statement: K.P. is the Chief Scientific Officer of Polgenix Inc. and a consultant of Prime Medicine and Editas Medicine. D.R.L. is a consultant and cofounder of Beam Therapeutics, Prime Medicine, Pairwise Plants, Editas Medicine, Chroma Medicine, and Nvelop Therapeutics, companies that use and/or deliver genome editing or genome engineering agents. The Broad Institute has filed patent applications on base editing.
Figures
References
-
- Georgiou M., Fujinami K., Michaelides M., Inherited retinal diseases: Therapeutics, clinical trials and end points—A review. Clin. Exp. Ophthalmol. 49, 270–288 (2021). - PubMed

