Natural Coevolution of Tumor and Immunoenvironment in Glioblastoma
- PMID: 36122307
- PMCID: PMC9716251
- DOI: 10.1158/2159-8290.CD-22-0196
Natural Coevolution of Tumor and Immunoenvironment in Glioblastoma
Abstract
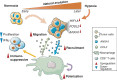
Isocitrate dehydrogenase (IDH) wild-type glioblastoma (GBM) has a dismal prognosis. A better understanding of tumor evolution holds the key to developing more effective treatment. Here we study GBM's natural evolutionary trajectory by using rare multifocal samples. We sequenced 61,062 single cells from eight multifocal IDH wild-type primary GBMs and defined a natural evolution signature (NES) of the tumor. We show that the NES significantly associates with the activation of transcription factors that regulate brain development, including MYBL2 and FOSL2. Hypoxia is involved in inducing NES transition potentially via activation of the HIF1A-FOSL2 axis. High-NES tumor cells could recruit and polarize bone marrow-derived macrophages through activation of the FOSL2-ANXA1-FPR1/3 axis. These polarized macrophages can efficiently suppress T-cell activity and accelerate NES transition in tumor cells. Moreover, the polarized macrophages could upregulate CCL2 to induce tumor cell migration.
Significance: GBM progression could be induced by hypoxia via the HIF1A-FOSL2 axis. Tumor-derived ANXA1 is associated with recruitment and polarization of bone marrow-derived macrophages to suppress the immunoenvironment. The polarized macrophages promote tumor cell NES transition and migration. This article is highlighted in the In This Issue feature, p. 2711.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures






Comment in
References
-
- Gatenby RA, Brown JS. Integrating evolutionary dynamics into cancer therapy. Nat Rev Clin Oncol 2020;17:675–86. - PubMed
-
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJB, Janzer RC, et al. . Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009;10:459–66. - PubMed
-
- Jiang T, Nam DH, Ram Z, Poon WS, Wang J, Boldbaatar D, et al. . Clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett 2021;499:60–72. - PubMed
-
- Mallick S, Benson R, Hakim A, Rath GK. Management of glioblastoma after recurrence: a changing paradigm. J Egypt Natl Canc Inst 2016;28:199–210. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

