Angiocrine extracellular vesicles impose mesenchymal reprogramming upon proneural glioma stem cells
- PMID: 36123372
- PMCID: PMC9485157
- DOI: 10.1038/s41467-022-33235-7
Angiocrine extracellular vesicles impose mesenchymal reprogramming upon proneural glioma stem cells
Abstract
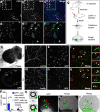
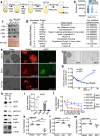
Glioblastoma (GBM) is an incurable form of primary astrocytic brain tumor driven by glioma stem cell (GSC) compartment closely associated with the vascular niche. GSC phenotypes are heterogeneous and range from proneural to mesenchymal-like, the latter characterised by greater invasiveness. Here we document the secretory (angiocrine) role of endothelial cells and their derived extracellular vesicles (EVs) as drivers of proneural-to-mesenchymal reprogramming of GSCs. These changes involve activation of matrix metalloproteinases (MMPs) and NFκB, and inactivation of NOTCH, while altering responsiveness to chemotherapy and driving infiltrative growth in the brain. Our findings suggest that EV-mediated angiocrine interactions impact the nature of cellular stemness in GBM with implications for disease biology and therapy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

