Clinically relevant pathogens on surfaces display differences in survival and transcriptomic response in relation to probiotic and traditional cleaning strategies
- PMID: 36123373
- PMCID: PMC9485146
- DOI: 10.1038/s41522-022-00335-7
Clinically relevant pathogens on surfaces display differences in survival and transcriptomic response in relation to probiotic and traditional cleaning strategies
Abstract
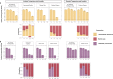
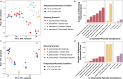
Indoor surfaces are paradoxically presumed to be both colonized by pathogens, necessitating disinfection, and "microbial wastelands." In these resource-poor, dry environments, competition and decay are thought to be important drivers of microbial community composition. However, the relative contributions of these two processes have not been specifically evaluated. To bridge this knowledge gap, we used microcosms to evaluate whether interspecies interactions occur on surfaces. We combined transcriptomics and traditional microbiology techniques to investigate whether competition occurred between two clinically important pathogens, Acinetobacter baumannii and Klebsiella pneumoniae, and a probiotic cleaner containing a consortium of Bacillus species. Probiotic cleaning seeks to take advantage of ecological principles such as competitive exclusion, thus using benign microorganisms to inhibit viable pathogens, but there is limited evidence that competitive exclusion in fact occurs in environments of interest (i.e., indoor surfaces). Our results indicate that competition in this setting has a negligible impact on community composition but may influence the functions expressed by active organisms. Although Bacillus spp. remained viable on surfaces for an extended period of time after application, viable colony forming units (CFUs) of A. baumannii recovered following exposure to a chemical-based detergent with and without Bacillus spp. showed no statistical difference. Similarly, for K. pneumoniae, there were small statistical differences in CFUs between cleaning scenarios with or without Bacillus spp. in the chemical-based detergent. The transcriptome of A. baumannii with and without Bacillus spp. exposure shared a high degree of similarity in overall gene expression, but the transcriptome of K. pneumoniae differed in overall gene expression, including reduced response in genes related to antimicrobial resistance. Together, these results highlight the need to fully understand the underlying biological and ecological mechanisms for community assembly and function on indoor surfaces, as well as having practical implications for cleaning and disinfection strategies for infection prevention.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Barber OW, Hartmann EM. Benzalkonium chloride: A systematic review of its environmental entry through wastewater treatment, potential impact, and mitigation strategies. Crit. Rev. Environ. Sci. Technol. 2021;0:1–30.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

