Asymmetric chromatin retention and nuclear envelopes separate chromosomes in fused cells in vivo
- PMID: 36123528
- PMCID: PMC9485224
- DOI: 10.1038/s42003-022-03874-z
Asymmetric chromatin retention and nuclear envelopes separate chromosomes in fused cells in vivo
Abstract
Hybrid cells derived through fertilization or somatic cell fusion recognize and separate chromosomes of different origins. The underlying mechanisms are unknown but could prevent aneuploidy and tumor formation. Here, we acutely induce fusion between Drosophila neural stem cells (neuroblasts; NBs) and differentiating ganglion mother cells (GMCs) in vivo to define how epigenetically distinct chromatin is recognized and segregated. We find that NB-GMC hybrid cells align both endogenous (neuroblast-origin) and ectopic (GMC-origin) chromosomes at the metaphase plate through centrosome derived dual-spindles. Physical separation of endogenous and ectopic chromatin is achieved through asymmetric, microtubule-dependent chromatin retention in interphase and physical boundaries imposed by nuclear envelopes. The chromatin separation mechanisms described here could apply to the first zygotic division in insects, arthropods, and vertebrates or potentially inform biased chromatid segregation in stem cells.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

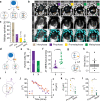


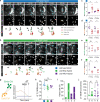
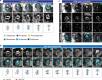

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

