Exploration of Hub Genes in Retinopathy of Prematurity Based on Bioinformatics Analysis of the Oxygen-Induced Retinopathy Model
- PMID: 36124139
- PMCID: PMC9482502
- DOI: 10.1155/2022/9835524
Exploration of Hub Genes in Retinopathy of Prematurity Based on Bioinformatics Analysis of the Oxygen-Induced Retinopathy Model
Abstract
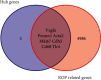
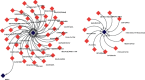
Retinopathy of prematurity (ROP) is a major blindness-causing disease that is characterized by an arrest of normal vascular development and neovascularization of the retina. Previous studies have shown that genetic factors may be associated with the development and severity of ROP. However, the genes and mechanisms underlying ROP remain unclear. We aimed to identify hub genes in ROP and drugs related to these genes by integrative analysis. The expression profiles of GSE158799 and GSE135844 were acquired from the Gene Expression Omnibus (GEO) database, and differentially expressed genes (DEGs) were identified. Then, an integrative analysis was performed including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), gene set enrichment analysis (GSEA), protein-protein interaction (PPI) network, transcription factor (TF)-gene, and miRNA-gene networks analysis. Moreover, we verified hub genes and identified potential drugs. 225 common DEGs were identified. Biological function analysis indicated that angiogenesis, cell surface, cell adhesion, extracellular matrix, and focal adhesion genes were enriched among DEGs. The PI3K/Akt signalingpathway, focal adhesion, and extracellular matrix (ECM)-receptor interaction were markedly enriched in the KEGG pathway analysis. Finally, 5 hub genes related to the nosogenesis of ROP were identified and found to be targeted by VEGFA inhibitors, TLR4 antagonists, and sunitinib. The present study showed that VEGFA, ACTA2, MKI67, CD68, and TLR4 are potential hub genes involved in the pathogenesis of ROP. Moreover, TLR4 antagonists and sunitinib may be new candidate drugs for ROP therapy, in addition to VEGFA inhibitors.
Copyright © 2022 Qi Xiong et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures







Similar articles
-
Identification of key candidate genes and biological pathways in bladder cancer.PeerJ. 2018 Dec 4;6:e6036. doi: 10.7717/peerj.6036. eCollection 2018. PeerJ. 2018. PMID: 30533316 Free PMC article.
-
Integrated bioinformatics analysis for the screening of hub genes and therapeutic drugs in ovarian cancer.J Ovarian Res. 2020 Jan 27;13(1):10. doi: 10.1186/s13048-020-0613-2. J Ovarian Res. 2020. PMID: 31987036 Free PMC article.
-
Identification of differentially expressed genes, associated functional terms pathways, and candidate diagnostic biomarkers in inflammatory bowel diseases by bioinformatics analysis.Exp Ther Med. 2019 Jul;18(1):278-288. doi: 10.3892/etm.2019.7541. Epub 2019 May 3. Exp Ther Med. 2019. PMID: 31258663 Free PMC article.
-
Molecular investigation of candidate genes for pyroptosis-induced inflammation in diabetic retinopathy.Front Endocrinol (Lausanne). 2022 Jul 25;13:918605. doi: 10.3389/fendo.2022.918605. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35957838 Free PMC article.
-
Exploration of the pathogenesis of Sjögren's syndrome via DNA methylation and transcriptome analyses.Clin Rheumatol. 2022 Sep;41(9):2765-2777. doi: 10.1007/s10067-022-06200-4. Epub 2022 May 13. Clin Rheumatol. 2022. PMID: 35562622
References
LinkOut - more resources
Full Text Sources
Miscellaneous

