Knowledge-guided deep learning models of drug toxicity improve interpretation
- PMID: 36124309
- PMCID: PMC9481960
- DOI: 10.1016/j.patter.2022.100565
Knowledge-guided deep learning models of drug toxicity improve interpretation
Abstract
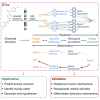
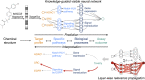
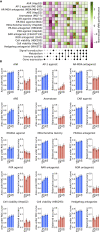
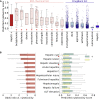
In drug development, a major reason for attrition is the lack of understanding of cellular mechanisms governing drug toxicity. The black-box nature of conventional classification models has limited their utility in identifying toxicity pathways. Here we developed DTox (deep learning for toxicology), an interpretation framework for knowledge-guided neural networks, which can predict compound response to toxicity assays and infer toxicity pathways of individual compounds. We demonstrate that DTox can achieve the same level of predictive performance as conventional models with a significant improvement in interpretability. Using DTox, we were able to rediscover mechanisms of transcription activation by three nuclear receptors, recapitulate cellular activities induced by aromatase inhibitors and pregnane X receptor (PXR) agonists, and differentiate distinctive mechanisms leading to HepG2 cytotoxicity. Virtual screening by DTox revealed that compounds with predicted cytotoxicity are at higher risk for clinical hepatic phenotypes. In summary, DTox provides a framework for deciphering cellular mechanisms of toxicity in silico.
Keywords: deep learning; drug toxicity; model interpretation; molecular toxicology.
© 2022 The Author(s).
Conflict of interest statement
J.H.M. is a member of the advisory board of Patterns.
Figures



Similar articles
-
Predicting drug toxicity at the intersection of informatics and biology: DTox builds a foundation.Patterns (N Y). 2022 Sep 9;3(9):100586. doi: 10.1016/j.patter.2022.100586. eCollection 2022 Sep 9. Patterns (N Y). 2022. PMID: 36124303 Free PMC article.
-
Revealing cytotoxic substructures in molecules using deep learning.J Comput Aided Mol Des. 2020 Jul;34(7):731-746. doi: 10.1007/s10822-020-00310-4. Epub 2020 Apr 16. J Comput Aided Mol Des. 2020. PMID: 32297073 Free PMC article.
-
Cytochrome P450 enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells.Toxicol In Vitro. 2007 Dec;21(8):1581-91. doi: 10.1016/j.tiv.2007.05.014. Epub 2007 Jun 8. Toxicol In Vitro. 2007. PMID: 17637504
-
A Review on the Recent Applications of Deep Learning in Predictive Drug Toxicological Studies.Chem Res Toxicol. 2023 Aug 21;36(8):1174-1205. doi: 10.1021/acs.chemrestox.2c00375. Epub 2023 Aug 10. Chem Res Toxicol. 2023. PMID: 37561655 Review.
-
Advancing Predictive Hepatotoxicity at the Intersection of Experimental, in Silico, and Artificial Intelligence Technologies.Chem Res Toxicol. 2018 Jun 18;31(6):412-430. doi: 10.1021/acs.chemrestox.8b00054. Epub 2018 May 10. Chem Res Toxicol. 2018. PMID: 29722533 Review.
Cited by
-
Reliable interpretability of biology-inspired deep neural networks.NPJ Syst Biol Appl. 2023 Oct 10;9(1):50. doi: 10.1038/s41540-023-00310-8. NPJ Syst Biol Appl. 2023. PMID: 37816807 Free PMC article.
-
Preclinical side effect prediction through pathway engineering of protein interaction network models.CPT Pharmacometrics Syst Pharmacol. 2024 Jul;13(7):1180-1200. doi: 10.1002/psp4.13150. Epub 2024 May 12. CPT Pharmacometrics Syst Pharmacol. 2024. PMID: 38736280 Free PMC article.
-
In silico off-target profiling for enhanced drug safety assessment.Acta Pharm Sin B. 2024 Jul;14(7):2927-2941. doi: 10.1016/j.apsb.2024.03.002. Epub 2024 Mar 6. Acta Pharm Sin B. 2024. PMID: 39027254 Free PMC article.
-
Predicting drug toxicity at the intersection of informatics and biology: DTox builds a foundation.Patterns (N Y). 2022 Sep 9;3(9):100586. doi: 10.1016/j.patter.2022.100586. eCollection 2022 Sep 9. Patterns (N Y). 2022. PMID: 36124303 Free PMC article.
-
Evaluating chemical effects on human neural cells through calcium imaging and deep learning.iScience. 2024 Nov 1;27(12):111298. doi: 10.1016/j.isci.2024.111298. eCollection 2024 Dec 20. iScience. 2024. PMID: 39634567 Free PMC article.
References
-
- Richard A.M., Huang R., Waidyanatha S., Shinn P., Collins B.J., Thillainadarajah I., Grulke C.M., Williams A.J., Lougee R.R., Judson R.S., et al. The Tox21 10K compound library: collaborative chemistry advancing toxicology. Chem. Res. Toxicol. 2021;34:189–216. doi: 10.1021/acs.chemrestox.0c00264. - DOI - PMC - PubMed
-
- Kleinstreuer N.C., Yang J., Berg E.L., Knudsen T.B., Richard A.M., Martin M.T., Reif D.M., Judson R.S., Polokoff M., Dix D.J., et al. Phenotypic screening of the ToxCast chemical library to classify toxic and therapeutic mechanisms. Nat. Biotechnol. 2014;32:583–591. doi: 10.1038/nbt.2914. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

