Protein metalation in a nutshell
- PMID: 36124565
- PMCID: PMC10087151
- DOI: 10.1002/1873-3468.14500
Protein metalation in a nutshell
Abstract
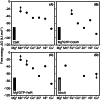
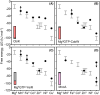
Metalation, the acquisition of metals by proteins, must avoid mis-metalation with tighter binding metals. This is illustrated by four selected proteins that require different metals: all show similar ranked orders of affinity for bioavailable metals, as described in a universal affinity series (the Irving-Williams series). Crucially, cellular protein metalation occurs in competition with other metal binding sites. The strength of this competition defines the intracellular availability of each metal: its magnitude has been estimated by calibrating a cells' set of DNA-binding, metal-sensing, transcriptional regulators. This has established that metal availabilities (as free energies for forming metal complexes) are maintained to the inverse of the universal series. The tightest binding metals are least available. With these availabilities, correct metalation is achieved.
Keywords: Irving-Williams series; cobalt; copper; iron; magnesium; manganese; metal sensor; metalation; nickel; zinc.
© 2022 The Authors. FEBS Letters published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Figures
References
-
- Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem. 2008;13(8):1205–18. - PubMed
-
- Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460(7257):823–30. - PubMed
-
- Tottey S, Waldron KJ, Firbank SJ, Reale B, Bessant C, Sato K, et al. Protein‐folding location can regulate manganese‐binding versus copper‐ or zinc‐binding. Nature. 2008;455(7216):1138–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

