QUADRatlas: the RNA G-quadruplex and RG4-binding proteins database
- PMID: 36124670
- PMCID: PMC9825518
- DOI: 10.1093/nar/gkac782
QUADRatlas: the RNA G-quadruplex and RG4-binding proteins database
Abstract
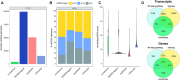
RNA G-quadruplexes (RG4s) are non-canonical, disease-associated post-transcriptional regulators of gene expression whose functions are driven by RNA-binding proteins (RBPs). Being able to explore transcriptome-wide RG4 formation and interaction with RBPs is thus paramount to understanding how they are regulated and exploiting them as potential therapeutic targets. Towards this goal, we present QUADRatlas (https://rg4db.cibio.unitn.it), a database of experimentally-derived and computationally predicted RG4s in the human transcriptome, enriched with biological function and disease associations. As RBPs are key to their function, we mined known interactions of RG4s with such proteins, complemented with an extensive RBP binding sites dataset. Users can thus intersect RG4s with their potential regulators and effectors, enabling the formulation of novel hypotheses on RG4 regulation, function and pathogenicity. To support this capability, we provide analysis tools for predicting whether an RBP can bind RG4s, RG4 enrichment in a gene set, and de novo RG4 prediction. Genome-browser and table views allow exploring, filtering, and downloading the data quickly for individual genes and in batch. QUADRatlas is a significant step forward in our ability to understand the biology of RG4s, offering unmatched data content and enabling the integrated analysis of RG4s and their interactions with RBPs.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Dumas L., Herviou P., Dassi E., Cammas A., Millevoi S.. G-Quadruplexes in RNA biology: recent advances and future directions. Trends Biochem. Sci. 2021; 46:270–283. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

