Biolink Model: A universal schema for knowledge graphs in clinical, biomedical, and translational science
- PMID: 36125173
- PMCID: PMC9372416
- DOI: 10.1111/cts.13302
Biolink Model: A universal schema for knowledge graphs in clinical, biomedical, and translational science
Abstract
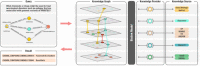
Within clinical, biomedical, and translational science, an increasing number of projects are adopting graphs for knowledge representation. Graph-based data models elucidate the interconnectedness among core biomedical concepts, enable data structures to be easily updated, and support intuitive queries, visualizations, and inference algorithms. However, knowledge discovery across these "knowledge graphs" (KGs) has remained difficult. Data set heterogeneity and complexity; the proliferation of ad hoc data formats; poor compliance with guidelines on findability, accessibility, interoperability, and reusability; and, in particular, the lack of a universally accepted, open-access model for standardization across biomedical KGs has left the task of reconciling data sources to downstream consumers. Biolink Model is an open-source data model that can be used to formalize the relationships between data structures in translational science. It incorporates object-oriented classification and graph-oriented features. The core of the model is a set of hierarchical, interconnected classes (or categories) and relationships between them (or predicates) representing biomedical entities such as gene, disease, chemical, anatomic structure, and phenotype. The model provides class and edge attributes and associations that guide how entities should relate to one another. Here, we highlight the need for a standardized data model for KGs, describe Biolink Model, and compare it with other models. We demonstrate the utility of Biolink Model in various initiatives, including the Biomedical Data Translator Consortium and the Monarch Initiative, and show how it has supported easier integration and interoperability of biomedical KGs, bringing together knowledge from multiple sources and helping to realize the goals of translational science.
© 2022 The Authors. Clinical and Translational Science published by Wiley Periodicals LLC on behalf of American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
The authors declared no competing interests for this work.
Figures

References
-
- Bisiani R, Shapiro SC. Encyclopedia of Artificial Intelligence. Beam search. Wiley; 1987.
-
- National Physical Laboratory . Symposium. International Conference on Machine Translation of Languages and Applied Language Analysis. National Physical Laboratory; 1961. Accessed March 03, 2022. https://market.android.com/details?id=book‐dTTawQEACAAJ
-
- Vrandečić D, Krötzsch M. Wikidata: A Free Collaborative Knowledge Base; 2014. Accessed March 03, 2022. https://ai.google/research/pubs/pub42240
-
- Vasilakes JA, Rizvi R, Zhang R. Annotated Semantic Predications from SemMedDB; 2018. Accessed March 03, 2022. https://conservancy.umn.edu/handle/11299/194965
Publication types
MeSH terms
Grants and funding
- OT3TR002019/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2TR003445/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2TR003449/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2TR002515/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2TR002584/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2TR003434/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2 TR003436/TR/NCATS NIH HHS/United States
- RM1 HG010860/HG/NHGRI NIH HHS/United States
- OT2TR003433/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2TR003435/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2TR002517/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT3TR002027/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2TR003422/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2 TR003434/TR/NCATS NIH HHS/United States
- OT2TR003441/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT3TR002020/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT3 TR002019/TR/NCATS NIH HHS/United States
- OT2TR003448/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2TR003428/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2TR002520/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2TR003427/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2TR003436/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2TR002514/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- R24 OD011883/OD/NIH HHS/United States
- OT2TR003443/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2 TR003443/TR/NCATS NIH HHS/United States
- OT3TR002025/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2 TR003428/TR/NCATS NIH HHS/United States
- OT2TR003437/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2TR003450/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT3TR002026/National Center for Advancing Translational Sciences, Biomedical Data Translator Program
- OT2 TR003422/TR/NCATS NIH HHS/United States
- OT2TR003430/National Center for Advancing Translational Sciences, Biomedical Data Translator Program

