Recycling of cell surface membrane proteins from yeast endosomes is regulated by ubiquitinated Ist1
- PMID: 36125415
- PMCID: PMC9491851
- DOI: 10.1083/jcb.202109137
Recycling of cell surface membrane proteins from yeast endosomes is regulated by ubiquitinated Ist1
Abstract
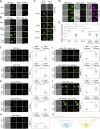
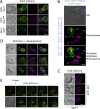
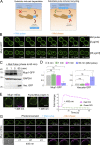
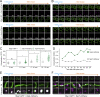
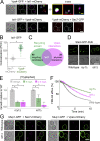
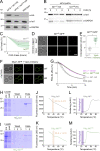
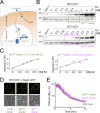
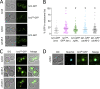
Upon internalization, many surface membrane proteins are recycled back to the plasma membrane. Although these endosomal trafficking pathways control surface protein activity, the precise regulatory features and division of labor between interconnected pathways are poorly defined. In yeast, we show recycling back to the surface occurs through distinct pathways. In addition to retrograde recycling pathways via the late Golgi, used by synaptobrevins and driven by cargo ubiquitination, we find nutrient transporter recycling bypasses the Golgi in a pathway driven by cargo deubiquitination. Nutrient transporters rapidly internalize to, and recycle from, endosomes marked by the ESCRT-III associated factor Ist1. This compartment serves as both "early" and "recycling" endosome. We show Ist1 is ubiquitinated and that this is required for proper endosomal recruitment and cargo recycling to the surface. Additionally, the essential ATPase Cdc48 and its adaptor Npl4 are required for recycling, potentially through regulation of ubiquitinated Ist1. This collectively suggests mechanistic features of recycling from endosomes to the plasma membrane are conserved.
© 2022 Laidlaw et al.
Figures





Comment in
-
A second chance at yeast early endosomes.J Cell Biol. 2022 Nov 7;221(11):e202210014. doi: 10.1083/jcb.202210014. Epub 2022 Oct 19. J Cell Biol. 2022. PMID: 36260296 Free PMC article.
References
-
- Adell, M.A.Y., Migliano S.M., Upadhyayula S., Bykov Y.S., Sprenger S., Pakdel M., Vogel G.F., Jih G., Skillern W., Behrouzi R., et al. . 2017. Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. Elife. 6:e31652. 10.7554/elife.31652 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

