Destabilization of mutated human PUS3 protein causes intellectual disability
- PMID: 36125428
- PMCID: PMC10092196
- DOI: 10.1002/humu.24471
Destabilization of mutated human PUS3 protein causes intellectual disability
Abstract
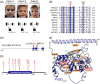
Pseudouridine (Ψ) is an RNA base modification ubiquitously found in many types of RNAs. In humans, the isomerization of uridine is catalyzed by different stand-alone pseudouridine synthases (PUS). Genomic mutations in the human pseudouridine synthase 3 gene (PUS3) have been identified in patients with neurodevelopmental disorders. However, the underlying molecular mechanisms that cause the disease phenotypes remain elusive. Here, we utilize exome sequencing to identify genomic variants that lead to a homozygous amino acid substitution (p.[(Tyr71Cys)];[(Tyr71Cys)]) in human PUS3 of two affected individuals and a compound heterozygous substitution (p.[(Tyr71Cys)];[(Ile299Thr)]) in a third patient. We obtain wild-type and mutated full-length human recombinant PUS3 proteins and characterize the enzymatic activity in vitro. Unexpectedly, we find that the p.Tyr71Cys substitution neither affect tRNA binding nor pseudouridylation activity in vitro, but strongly impair the thermostability profile of PUS3, while the p.Ile299Thr mutation causes protein aggregation. Concomitantly, we observe that the PUS3 protein levels as well as the level of PUS3-dependent Ψ levels are strongly reduced in fibroblasts derived from all three patients. In summary, our results directly illustrate the link between the identified PUS3 variants and reduced Ψ levels in the patient cells, providing a molecular explanation for the observed clinical phenotypes.
Keywords: PUS3; intellectual disorder; protein stability; pseudouridylation; tRNA modification.
© 2022 The Authors. Human Mutation published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Angelova, M. T. , Dimitrova, D. G. , Dinges, N. , Lence, T. , Worpenberg, L. , Carré, C. , & Roignant, J. Y. (2018). The emerging field of epitranscriptomics in neurodevelopmental and neuronal disorders. Frontiers in Bioengineering and Biotechnology, 6, 46. 10.3389/fbioe.2018.00046 - DOI - PMC - PubMed
-
- Begik, O. , Lucas, M. C. , Pryszcz, L. P. , Ramirez, J. M. , Medina, R. , Milenkovic, I. , Cruciani, S. , Liu H., Vieira H. G. S., Sas‐Chen A., Mattick J. S., Schwartz S., Novoa E. M. (2021). Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nature Biotechnology, 39(10), 1278–1291. 10.1038/s41587-021-00915-6 - DOI - PubMed
-
- Boccaletto, P. , MacHnicka, M. A. , Purta, E. , Piątkowski, P. , Bagiński, B. , Wirecki, T. K. , De crécy‐Lagard, V. , Ross, R. , Limbach, P. A. , Kotter, A. , Helm, M. , & Bujnicki, J. M. (2018). MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Research, 46(D1), D303–D307. 10.1093/nar/gkx1030 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

