A computational approach to biological pathogenicity
- PMID: 36125534
- PMCID: PMC9486766
- DOI: 10.1007/s00438-022-01951-w
A computational approach to biological pathogenicity
Abstract
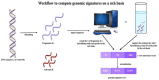
The current pandemic (COVID-19) has made evident the need to approach pathogenicity from a deeper and more systematic perspective that might lead to methodologies to quickly predict new strains of microbes that could be pathogenic to humans. Here we propose as a solution a general and principled definition of pathogenicity that can be practically implemented in operational ways in a framework for characterizing and assessing the (degree of) potential pathogenicity of a microbe to a given host (e.g., a human individual) just based on DNA biomarkers, and to the point of predicting its impact on a host a priori to a meaningful degree of accuracy. The definition is based on basic biochemistry, the Gibbs free Energy of duplex formation between oligonucleotides and some deep structural properties of DNA revealed by an approximation with certain properties. We propose two operational tests based on the nearest neighbor (NN) model of the Gibbs Energy and an approximating metric (the h-distance.) Quality assessments demonstrate that these tests predict pathogenicity with an accuracy of over 80%, and sensitivity and specificity over 90%. Other tests obtained by training machine learning models on deep features extracted from DNA sequences yield scores of 90% for accuracy, 100% for sensitivity and 80% for specificity. These results hint towards the possibility of an operational, objective, and general conceptual framework for prior identification of pathogens and their impact without the cost of death or sickness in a host (e.g., humans.) Consequently, a reasonable prediction of possible pathogens might pave the way to eventually transform the way we handle and prepare for future pandemic events and mitigate the adverse impact on human health, while reducing the number of clinical trials to obtain similar results.
Keywords: Digital genomic signature; Gibbs energy; Hybridization; Machine learning; Pathogenic relationship; Pathogens/nonpathogens; h-distance.
© 2022. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002) Introduction to pathogens. In Molecular biology of the cell, 4th edn. Garland Science.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

