Blockade of TGF-β signaling reactivates HIV-1/SIV reservoirs and immune responses in vivo
- PMID: 36125890
- PMCID: PMC9675457
- DOI: 10.1172/jci.insight.162290
Blockade of TGF-β signaling reactivates HIV-1/SIV reservoirs and immune responses in vivo
Erratum in
-
Blockade of TGF-β signaling reactivates HIV-1/SIV reservoirs and immune responses in vivo.JCI Insight. 2023 Nov 22;8(22):e176882. doi: 10.1172/jci.insight.176882. JCI Insight. 2023. PMID: 37991025 Free PMC article. No abstract available.
Abstract
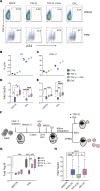
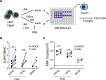
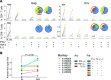
TGF-β plays a critical role in maintaining immune cells in a resting state by inhibiting cell activation and proliferation. Resting HIV-1 target cells represent the main cellular reservoir after long-term antiretroviral therapy (ART). We hypothesized that releasing cells from TGF-β-driven signaling would promote latency reversal. To test our hypothesis, we compared HIV-1 latency models with and without TGF-β and a TGF-β type 1 receptor inhibitor, galunisertib. We tested the effect of galunisertib in SIV-infected, ART-treated macaques by monitoring SIV-env expression via PET/CT using the 64Cu-DOTA-F(ab')2 p7D3 probe, along with plasma and tissue viral loads (VLs). Exogenous TGF-β reduced HIV-1 reactivation in U1 and ACH-2 models. Galunisertib increased HIV-1 latency reversal ex vivo and in PBMCs from HIV-1-infected, ART-treated, aviremic donors. In vivo, oral galunisertib promoted increased total standardized uptake values in PET/CT images in gut and lymph nodes of 5 out of 7 aviremic, long-term ART-treated, SIV-infected macaques. This increase correlated with an increase in SIV RNA in the gut. Two of the 7 animals also exhibited increases in plasma VLs. Higher anti-SIV T cell responses and antibody titers were detected after galunisertib treatment. In summary, our data suggest that blocking TGF-β signaling simultaneously increases retroviral reactivation events and enhances anti-SIV immune responses.
Keywords: AIDS/HIV; Cytokines; Immunotherapy; T cells.
Figures


Similar articles
-
TGF-β blockade drives a transitional effector phenotype in T cells reversing SIV latency and decreasing SIV reservoirs in vivo.Nat Commun. 2024 Feb 14;15(1):1348. doi: 10.1038/s41467-024-45555-x. Nat Commun. 2024. PMID: 38355731 Free PMC article.
-
Brain Macrophages in Simian Immunodeficiency Virus-Infected, Antiretroviral-Suppressed Macaques: a Functional Latent Reservoir.mBio. 2017 Aug 15;8(4):e01186-17. doi: 10.1128/mBio.01186-17. mBio. 2017. PMID: 28811349 Free PMC article.
-
TGF-β blockade drives a transitional effector phenotype in T cells reversing SIV latency and decreasing SIV reservoirs in vivo.bioRxiv [Preprint]. 2023 Dec 15:2023.09.05.556422. doi: 10.1101/2023.09.05.556422. bioRxiv. 2023. Update in: Nat Commun. 2024 Feb 14;15(1):1348. doi: 10.1038/s41467-024-45555-x. PMID: 38014094 Free PMC article. Updated. Preprint.
-
Brain macrophages harbor latent, infectious simian immunodeficiency virus.AIDS. 2019 Dec 1;33 Suppl 2(Suppl 2):S181-S188. doi: 10.1097/QAD.0000000000002269. AIDS. 2019. PMID: 31789817 Free PMC article. Review.
-
Mechanisms of CD8+ T cell-mediated suppression of HIV/SIV replication.Eur J Immunol. 2018 Jun;48(6):898-914. doi: 10.1002/eji.201747172. Epub 2018 Mar 26. Eur J Immunol. 2018. PMID: 29427516 Free PMC article. Review.
Cited by
-
Whole-body PET imaging of simian immunodeficiency virus using gp120-targeting probes fails to reveal regions of specific uptake in rhesus macaques.Eur J Nucl Med Mol Imaging. 2025 Jun;52(7):2645-2657. doi: 10.1007/s00259-025-07110-8. Epub 2025 Jan 31. Eur J Nucl Med Mol Imaging. 2025. PMID: 39888424 Free PMC article.
-
Challenges in HIV-1 Latent Reservoir and Target Cell Quantification in CAR-T Cell and Other Lentiviral Gene Modifying HIV Cure Strategies.Viruses. 2023 May 9;15(5):1126. doi: 10.3390/v15051126. Viruses. 2023. PMID: 37243212 Free PMC article. Review.
-
HIV-SEQ REVEALS GLOBAL HOST GENE EXPRESSION DIFFERENCES BETWEEN HIV-TRANSCRIBING CELLS FROM VIREMIC AND SUPPRESSED PEOPLE WITH HIV.bioRxiv [Preprint]. 2024 Dec 20:2024.12.17.629023. doi: 10.1101/2024.12.17.629023. bioRxiv. 2024. PMID: 39763963 Free PMC article. Preprint.
-
Footprints of innate immune activity during HIV-1 reservoir cell evolution in early-treated infection.J Exp Med. 2024 Nov 4;221(11):e20241091. doi: 10.1084/jem.20241091. Epub 2024 Oct 28. J Exp Med. 2024. PMID: 39466203 Free PMC article.
-
Polymorphonuclear myeloid-derived suppressor cells regulates immune recovery during HIV infection through PD-L1 and TGF-β pathways.Front Cell Infect Microbiol. 2024 Dec 17;14:1516421. doi: 10.3389/fcimb.2024.1516421. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39742336 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

