A MIR17HG-derived long noncoding RNA provides an essential chromatin scaffold for protein interaction and myeloma growth
- PMID: 36126301
- PMCID: PMC10082365
- DOI: 10.1182/blood.2022016892
A MIR17HG-derived long noncoding RNA provides an essential chromatin scaffold for protein interaction and myeloma growth
Abstract
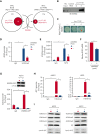
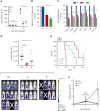
Long noncoding RNAs (lncRNAs) can drive tumorigenesis and are susceptible to therapeutic intervention. Here, we used a large-scale CRISPR interference viability screen to interrogate cell-growth dependency to lncRNA genes in multiple myeloma (MM) and identified a prominent role for the miR-17-92 cluster host gene (MIR17HG). We show that an MIR17HG-derived lncRNA, named lnc-17-92, is the main mediator of cell-growth dependency acting in a microRNA- and DROSHA-independent manner. Lnc-17-92 provides a chromatin scaffold for the functional interaction between c-MYC and WDR82, thus promoting the expression of ACACA, which encodes the rate-limiting enzyme of de novo lipogenesis acetyl-coA carboxylase 1. Targeting MIR17HG pre-RNA with clinically applicable antisense molecules disrupts the transcriptional and functional activities of lnc-17-92, causing potent antitumor effects both in vitro and in vivo in 3 preclinical animal models, including a clinically relevant patient-derived xenograft NSG mouse model. This study establishes a novel oncogenic function of MIR17HG and provides potent inhibitors for translation to clinical trials.
© 2023 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: N.C.M. serves on advisory boards of and as consultant to Takeda, BMS, Celgene, Janssen, Amgen, AbbVie, Oncopep, Karyopharm, Adaptive Biotechnology, and Novartis and holds equity ownership in Oncopep. K.C.A. serves on advisory boards to Janssen, Pfizer, Astrazeneca, Amgen, Precision Biosciences, Mana, Starton, and Raqia and is a Scientific Founder of OncoPep and C4 Therapeutics. R.A.Y. is a founder and shareholder of Syros Pharmaceuticals, Camp4 Therapeutics, Omega Therapeutics, and Dewpoint Therapeutics. E.M., S.G., and N.C.M filed a provisional patent on
Figures

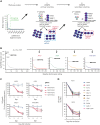
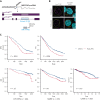

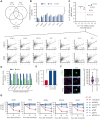
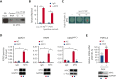
Comment in
-
RROL lncRNA role in multiple myeloma.Blood. 2023 Jan 26;141(4):328-330. doi: 10.1182/blood.2022018471. Blood. 2023. PMID: 36701172 No abstract available.
References
-
- Pichiorri F, Suh SS, Rocci A, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2016;30(2):349–351. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

