Ginsenoside Rd ameliorates muscle wasting by suppressing the signal transducer and activator of transcription 3 pathway
- PMID: 36127129
- PMCID: PMC9745546
- DOI: 10.1002/jcsm.13084
Ginsenoside Rd ameliorates muscle wasting by suppressing the signal transducer and activator of transcription 3 pathway
Abstract
Background: The effects of some drugs, aging, cancers, and other diseases can cause muscle wasting. Currently, there are no effective drugs for treating muscle wasting. In this study, the effects of ginsenoside Rd (GRd) on muscle wasting were studied.
Methods: Tumour necrosis factor-alpha (TNF-α)/interferon-gamma (IFN-γ)-induced myotube atrophy in mouse C2C12 and human skeletal myoblasts (HSkM) was evaluated based on cell thickness. Atrophy-related signalling, reactive oxygen species (ROS) level, mitochondrial membrane potential, and mitochondrial number were assessed. GRd (10 mg/kg body weight) was orally administered to aged mice (23-24 months old) and tumour-bearing (Lewis lung carcinoma [LLC1] or CT26) mice for 5 weeks and 16 days, respectively. Body weight, grip strength, inverted hanging time, and muscle weight were assessed. Histological analysis was also performed to assess the effects of GRd. The evolutionary chemical binding similarity (ECBS) approach, molecular docking, Biacore assay, and signal transducer and activator of transcription (STAT) 3 reporter assay were used to identify targets of GRd.
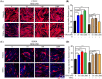
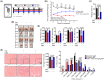
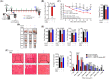
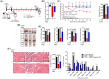
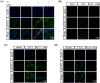
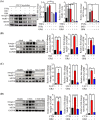
Results: GRd significantly induced hypertrophy in the C2C12 and HSkM myotubes (average diameter 50.8 ± 2.6% and 49.9% ± 3.7% higher at 100 nM, vs. control, P ≤ 0.001). GRd treatment ameliorated aging- and cancer-induced (LLC1 or CT26) muscle atrophy in mice, which was evidenced by significant increases in grip strength, hanging time, muscle mass, and muscle tissue cross-sectional area (1.3-fold to 4.6-fold, vs. vehicle, P ≤ 0.05; P ≤ 0.01; P ≤ 0.001). STAT3 was found to be a possible target of GRd by the ECBS approach and molecular docking assay. Validation of direct interaction between GRd and STAT3 was confirmed through Biacore analysis. GRd also inhibited STAT3 phosphorylation and STAT3 reporter activity, which led to the inhibition of STAT3 nuclear translocation and the suppression of downstream targets of STAT3, such as atrogin-1, muscle-specific RING finger protein (MuRF-1), and myostatin (MSTN) (29.0 ± 11.2% to 84.3 ± 30.5%, vs. vehicle, P ≤ 0.05; P ≤ 0.01; P ≤ 0.001). Additionally, GRd scavenged ROS (91.7 ± 1.4% reduction at 1 nM, vs. vehicle, P ≤ 0.001), inhibited TNF-α-induced dysregulation of ROS level, and improved mitochondrial integrity (P ≤ 0.05; P ≤ 0.01; P ≤ 0.001).
Conclusions: GRd ameliorates aging- and cancer-induced muscle wasting. Our findings suggest that GRd may be a novel therapeutic agent or adjuvant for reversing muscle wasting.
Keywords: cachexia; ginsenoside Rd; muscle wasting; sarcopenia; signal transducer and activator of transcription 3.
© 2022 The Authors. Journal of Cachexia, Sarcopenia and Muscle published by John Wiley & Sons Ltd on behalf of Society on Sarcopenia, Cachexia and Wasting Disorders.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Cruz‐Jentoft AJ, Sayer AA. Sarcopenia. Lancet 2019;393:2636–2646. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

