Deciphering multi-way interactions in the human genome
- PMID: 36127324
- PMCID: PMC9489732
- DOI: 10.1038/s41467-022-32980-z
Deciphering multi-way interactions in the human genome
Abstract
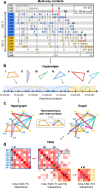
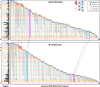
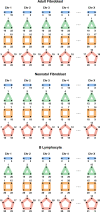
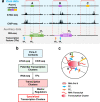
Chromatin architecture, a key regulator of gene expression, can be inferred using chromatin contact data from chromosome conformation capture, or Hi-C. However, classical Hi-C does not preserve multi-way contacts. Here we use long sequencing reads to map genome-wide multi-way contacts and investigate higher order chromatin organization in the human genome. We use hypergraph theory for data representation and analysis, and quantify higher order structures in neonatal fibroblasts, biopsied adult fibroblasts, and B lymphocytes. By integrating multi-way contacts with chromatin accessibility, gene expression, and transcription factor binding, we introduce a data-driven method to identify cell type-specific transcription clusters. We provide transcription factor-mediated functional building blocks for cell identity that serve as a global signature for cell types.
© 2022. The Author(s).
Conflict of interest statement
S.D. is an employee of iReprogram, L.L.C. L.M. and I.R. are co-founders of iReprogram, L.L.C. N.B. is an employee of Oxford Nanopore Technologies. S.L., C.C., and I.R. have submitted a patent application for the computational framework (2115-008250-US-PS1). The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

