Single cell atlas identifies lipid-processing and immunomodulatory endothelial cells in healthy and malignant breast
- PMID: 36127427
- PMCID: PMC9489707
- DOI: 10.1038/s41467-022-33052-y
Single cell atlas identifies lipid-processing and immunomodulatory endothelial cells in healthy and malignant breast
Abstract
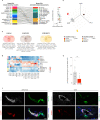
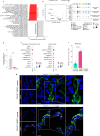
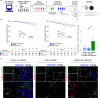
Since a detailed inventory of endothelial cell (EC) heterogeneity in breast cancer (BC) is lacking, here we perform single cell RNA-sequencing of 26,515 cells (including 8433 ECs) from 9 BC patients and compare them to published EC taxonomies from lung tumors. Angiogenic ECs are phenotypically similar, while other EC subtypes are different. Predictive interactome analysis reveals known but also previously unreported receptor-ligand interactions between ECs and immune cells, suggesting an involvement of breast EC subtypes in immune responses. We also identify a capillary EC subtype (LIPEC (Lipid Processing EC)), which expresses genes involved in lipid processing that are regulated by PPAR-γ and is more abundant in peri-tumoral breast tissue. Retrospective analysis of 4648 BC patients reveals that treatment with metformin (an indirect PPAR-γ signaling activator) provides long-lasting clinical benefit and is positively associated with LIPEC abundance. Our findings warrant further exploration of this LIPEC/PPAR-γ link for BC treatment.
© 2022. The Author(s).
Conflict of interest statement
K.P. is a speaker and/or consultant for Astra Zeneca, Eli Lilly, Exact Sciences, Focus Patient, Gilead Sciences, Medscape/Genomic Health, MSD, Novartis, Pfizer, Roche, Seagen (outside the submitted work), and has received research/travel support from Sanofi, MSD, Astra Zeneca, Novartis, Pfizer, PharmaMar, Roche (outside the submitted work). The other authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

