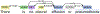Radiology Text Analysis System (RadText): Architecture and Evaluation
- PMID: 36128510
- PMCID: PMC9484781
- DOI: 10.1109/ichi54592.2022.00050
Radiology Text Analysis System (RadText): Architecture and Evaluation
Abstract
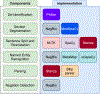
Analyzing radiology reports is a time-consuming and error-prone task, which raises the need for an efficient automated radiology report analysis system to alleviate the workloads of radiologists and encourage precise diagnosis. In this work, we present RadText, a high-performance open-source Python radiology text analysis system. RadText offers an easy-to-use text analysis pipeline, including de-identification, section segmentation, sentence split and word tokenization, named entity recognition, parsing, and negation detection. Superior to existing widely used toolkits, RadText features a hybrid text processing schema, supports raw text processing and local processing, which enables higher accuracy, better usability and improved data privacy. RadText adopts BioC as the unified interface, and also standardizes the output into a structured representation that is compatible with Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM), which allows for a more systematic approach to observational research across multiple, disparate data sources. We evaluated RadText on the MIMIC-CXR dataset, with five new disease labels that we annotated for this work. RadText demonstrates highly accurate classification performances, with a 0.91 average precision, 0.94 average recall and 0.92 average F-1 score. We also annotated a test set for the five new disease labels to facilitate future research or applications. We have made our code, documentations, examples and the test set available at https://github.com/bionlplab/radtext.
Keywords: Natural Language Processing; Radiology; Text Analysis Systems.
Figures
References
-
- Savova G, Masanz J, Ogren P, Zheng J, Sohn S, Kipper-Schuler K, and Chute C, “Mayo clinical text analysis and knowledge extraction system (ctakes): Architecture, component evaluation and applications,” Journal of the American Medical Informatics Association : JAMIA, vol. 17, pp. 507–13, 09 2010. - PMC - PubMed
-
- Neumann M, King D, Beltagy I, and Ammar W, “ScispaCy: Fast and Robust Models for Biomedical Natural Language Processing,” in Proceedings of the 18th BioNLP Workshop and Shared Task. Florence, Italy: Association for Computational Linguistics, Aug. 2019, pp. 319–327. [Online]. Available: https://www.aclweb.org/anthology/W19-5034
-
- Liu H, Bielinski SJ, Sohn S, Murphy S, Wagholikar KB, Jonnalagadda SR, Ravikumar K, Wu ST, Kullo IJ, and Chute CG, “An information extraction framework for cohort identification using electronic health records,” AMIA Joint Summits on Translational Science proceedings. AMIA Joint Summits on Translational Science, vol. 2013, p. 149–153, 2013. [Online]. Available: https://europepmc.org/articles/PMC3845757 - PMC - PubMed
-
- Soysal E, Wang J, Jiang M, Wu Y, Pakhomov S, Liu H, and Xu H, “CLAMP – a toolkit for efficiently building customized clinical natural language processing pipelines,” Journal of the American Medical Informatics Association, vol. 25, no. 3, pp. 331–336, 11 2017. [Online]. Available: 10.1093/jamia/ocx132 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous