Genome size and endoreplication in two pairs of cytogenetically contrasting species of Pulmonaria (Boraginaceae) in Central Europe
- PMID: 36128515
- PMCID: PMC9476981
- DOI: 10.1093/aobpla/plac036
Genome size and endoreplication in two pairs of cytogenetically contrasting species of Pulmonaria (Boraginaceae) in Central Europe
Abstract
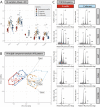
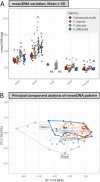
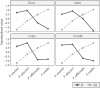
Genome size is species-specific feature and commonly constant in an organism. In various plants, DNA content in cell nucleus is commonly increased in process of endoreplication, cellular-specific multiplication of DNA content without mitosis. This leads to the endopolyploidy, the presence of multiplied chromosome sets in a subset of cells. The relationship of endopolyploidy to species-specific genome size is rarely analysed and is not fully understood. While negative correlation between genome size and endopolyploidy level is supposed, this is species- and lineage-specific. In the present study, we shed light on this topic, exploring both genome size and endoreplication-induced DNA content variation in two pairs of morphologically similar species of Pulmonaria, P. obscura-P. officinalis and P. mollis-P. murinii. We aim (i) to characterize genome size and chromosome numbers in these species using cytogenetic, root-tip squashing and flow cytometry (FCM) techniques; (ii) to investigate the degree of endopolyploidy in various plant organs, including the root, stem, leaf, calyx and corolla using FCM; and (iii) to comprehensively characterize and compare the level of endopolyploidy and DNA content in various organs of all four species in relation to species systematic relationships and genome size variation. We have confirmed the diploid-dysploid nature of chromosome complements, and divergent genome sizes for Pulmonaria species: P. murinii with 2n = 2x = 14, 2.31 pg/2C, P. obscura 2n = 2x = 14, 2.69 pg/2C, P. officinalis 2n = 2x = 16, 2.96 pg/2C and P. mollis 2n = 2x = 18, 3.18 pg/2C. Endopolyploidy varies between species and organs, and we have documented 4C-8C in all four organs and up to 32C (64C) endopolyploid nuclei in stems at least in some species. Two species with lower genome sizes tend to have higher endopolyploidy levels than their closest relatives. Endoreplication-generated tissue-specific mean DNA content is increased and more balanced among species in all four organs compared to genome size. Our results argue for the narrow relationship between genome size and endopolyploidy in the present plant group within the genus Pulmonaria, and endopolyploidization seems to play a compensatory developmental role in organs of related morphologically similar species.
Keywords: Boraginaceae; Pulmonaria; endoreplication; flow cytometry; genome size; geophytes.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Annals of Botany Company.
Figures

Similar articles
-
First insight into the genomes of the Pulmonaria officinalis group (Boraginaceae) provided by repeatome analysis and comparative karyotyping.BMC Plant Biol. 2024 Sep 13;24(1):859. doi: 10.1186/s12870-024-05497-4. BMC Plant Biol. 2024. PMID: 39266954 Free PMC article.
-
Chromosome Number, Ploidy Level, and Nuclear DNA Content in 23 Species of Echeveria (Crassulaceae).Genes (Basel). 2021 Dec 3;12(12):1950. doi: 10.3390/genes12121950. Genes (Basel). 2021. PMID: 34946899 Free PMC article.
-
Systemic endopolyploidy in Spathoglottis plicata (Orchidaceae) development.BMC Cell Biol. 2004 Sep 1;5:33. doi: 10.1186/1471-2121-5-33. BMC Cell Biol. 2004. PMID: 15341672 Free PMC article.
-
Endopolyploidy as a potential driver of animal ecology and evolution.Biol Rev Camb Philos Soc. 2017 Feb;92(1):234-247. doi: 10.1111/brv.12226. Epub 2015 Oct 14. Biol Rev Camb Philos Soc. 2017. PMID: 26467853 Review.
-
Endopolyploidy in seed plants.Bioessays. 2006 Mar;28(3):271-81. doi: 10.1002/bies.20371. Bioessays. 2006. PMID: 16479577 Review.
Cited by
-
Mixed-Ploidy and Dysploidy in Hypericum perforatum: A Karyomorphological and Genome Size Study.Plants (Basel). 2022 Nov 12;11(22):3068. doi: 10.3390/plants11223068. Plants (Basel). 2022. PMID: 36432797 Free PMC article.
-
First insight into the genomes of the Pulmonaria officinalis group (Boraginaceae) provided by repeatome analysis and comparative karyotyping.BMC Plant Biol. 2024 Sep 13;24(1):859. doi: 10.1186/s12870-024-05497-4. BMC Plant Biol. 2024. PMID: 39266954 Free PMC article.
-
Endoreplication-Why Are We Not Using Its Full Application Potential?Int J Mol Sci. 2023 Jul 24;24(14):11859. doi: 10.3390/ijms241411859. Int J Mol Sci. 2023. PMID: 37511616 Free PMC article. Review.
References
-
- Bainard JD, Bainard LD, Henry TA, Fazekas AJ, Newmaster SG.. 2012. A multivariate analysis of variation in genome size and endoreduplication in angiosperms reveals strong phylogenetic signal and association with phenotypic traits. New Phytologist 196:1240–1250. - PubMed
-
- Bainard LD, Bainard JD, Newmaster SG, Klironomos JN.. 2011. Mycorrhizal symbiosis stimulates endoreduplication in angiosperms. Plant, Cell and Environment 34:1577–1585. - PubMed
-
- Barow M. 2006. Endopolyploidy in seed plants. Bioessays 28:271–281. - PubMed
LinkOut - more resources
Full Text Sources

