Benzodioxole grafted spirooxindole pyrrolidinyl derivatives: synthesis, characterization, molecular docking and anti-diabetic activity
- PMID: 36128541
- PMCID: PMC9404121
- DOI: 10.1039/d2ra04452h
Benzodioxole grafted spirooxindole pyrrolidinyl derivatives: synthesis, characterization, molecular docking and anti-diabetic activity
Abstract
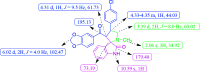
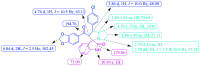
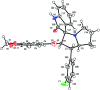
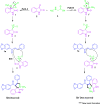
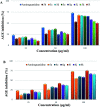
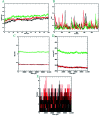
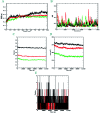
A highly stereoselective, three-component method has been developed to synthesize pyrrolidine and pyrrolizidine containing spirooxindole derivatives. The interaction between the dipolarophile α,β-unsaturated carbonyl compounds and the dipole azomethine ylide formed in situ by the reaction of 1,2-dicarbonyl compounds and secondary amino acids is referred to as the 1,3-dipolar cycloaddition reaction. The reaction conditions were optimized to achieve excellent stereo- and regioselectivity. Shorter reaction time, simple work-up and excellent yields are the salient features of the present approach. Various spectroscopic methods and single crystal X-ray diffraction examinations of one example of compound 6i validated the stereochemistry of the expected products. The anti-diabetic activity of the newly synthesized spirooxindole derivatives was tested against the α-glucosidase and α-amylase enzymes. Compound 6i was found to exhibit potent inhibition activity against α-glucosidase and α-amylase enzymes which is further evidenced by molecular docking studies.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



Similar articles
-
Stereoselective Synthesis of Spirooxindole Derivatives Using One-Pot Multicomponent Cycloaddition Reaction and Evaluation of Their Antiproliferative Efficacy.ACS Omega. 2020 Oct 16;5(42):27332-27343. doi: 10.1021/acsomega.0c03675. eCollection 2020 Oct 27. ACS Omega. 2020. PMID: 33134696 Free PMC article.
-
Combinatorial synthesis of functionalized spirooxindole-pyrrolidine/pyrrolizidine/pyrrolothiazole derivatives via three-component 1,3-dipolar cycloaddition reactions.ACS Comb Sci. 2014 Sep 8;16(9):506-12. doi: 10.1021/co500085t. Epub 2014 Jul 29. ACS Comb Sci. 2014. PMID: 25033950
-
Regio- and stereoselective synthesis of new spirooxindoles via 1,3-dipolar cycloaddition reaction: Anticancer and molecular docking studies.J Photochem Photobiol B. 2018 Mar;180:98-108. doi: 10.1016/j.jphotobiol.2018.01.026. Epub 2018 Jan 31. J Photochem Photobiol B. 2018. PMID: 29413708
-
Pyrano[3,2-c]quinoline Derivatives as New Class of α-glucosidase Inhibitors to Treat Type 2 Diabetes: Synthesis, in vitro Biological Evaluation and Kinetic Study.Med Chem. 2019;15(1):8-16. doi: 10.2174/1573406414666180528110104. Med Chem. 2019. PMID: 29807519 Review.
-
Diastereodivergent Asymmetric 1,3-Dipolar Cycloaddition of Azomethine Ylides and β-Fluoroalkyl Vinylsulfones: Low Copper(II) Catalyst Loading and Theoretical Studies.Angew Chem Int Ed Engl. 2019 Nov 11;58(46):16637-16643. doi: 10.1002/anie.201908227. Epub 2019 Sep 30. Angew Chem Int Ed Engl. 2019. PMID: 31482632 Review.
Cited by
-
Synthesis of Tetracyclic Spirooxindolepyrrolidine-Engrafted Hydantoin Scaffolds: Crystallographic Analysis, Molecular Docking Studies and Evaluation of Their Antimicrobial, Anti-Inflammatory and Analgesic Activities.Molecules. 2023 Nov 6;28(21):7443. doi: 10.3390/molecules28217443. Molecules. 2023. PMID: 37959862 Free PMC article.
-
Decarboxylative 1,3-dipolar cycloadditions of l-proline.RSC Adv. 2024 Mar 13;14(12):8481-8501. doi: 10.1039/d3ra08160e. eCollection 2024 Mar 6. RSC Adv. 2024. PMID: 38482067 Free PMC article. Review.
-
Dipolarophile-Controlled Regioselective 1,3-Dipolar Cycloaddition: A Switchable Divergent Access to Functionalized N-Fused Pyrrolidinyl Spirooxindoles.Int J Mol Sci. 2023 Feb 13;24(4):3771. doi: 10.3390/ijms24043771. Int J Mol Sci. 2023. PMID: 36835183 Free PMC article.
-
Antidiabetic Activity of Potential Probiotics Limosilactobacillus spp., Levilactobacillus spp., and Lacticaseibacillus spp. Isolated from Fermented Sugarcane Juice: A Comprehensive In Vitro and In Silico Study.Nutrients. 2023 Apr 13;15(8):1882. doi: 10.3390/nu15081882. Nutrients. 2023. PMID: 37111101 Free PMC article.
-
Highly efficient, catalyst-free, one-pot sequential four-component synthesis of novel spiroindolinone-pyrazole scaffolds as anti-Alzheimer agents: in silico study and biological screening.RSC Med Chem. 2023 Dec 4;15(1):207-222. doi: 10.1039/d3md00255a. eCollection 2024 Jan 25. RSC Med Chem. 2023. PMID: 38283217 Free PMC article.
References
-
- Marti C. Careira E. M. Eur. J. Med. Chem. 2003;12:2209–2219.
-
- Teja C. Babu S. N. Noor A. Daniel J. A. Devi S. A. Rahman F. Khan N. RSC Adv. 2020;10:12262–12271. doi: 10.1039/D0RA01553A. - DOI - PMC - PubMed
- Toumi A. Boudriga S. Hamden K. Sobeh M. Cheurfa M. Askri M. Knorr M. Strohmann C. Brieger L. Bioorg. Chem. 2021;106:104507. doi: 10.1016/j.bioorg.2020.104507. - DOI - PubMed
LinkOut - more resources
Full Text Sources

